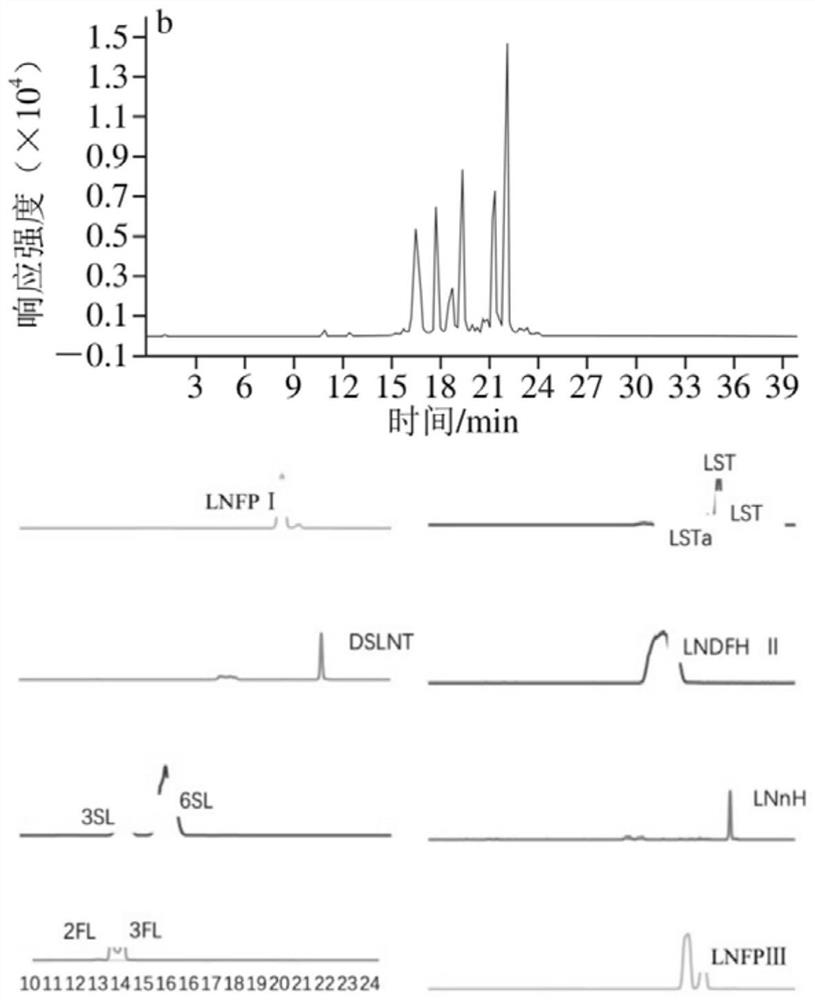
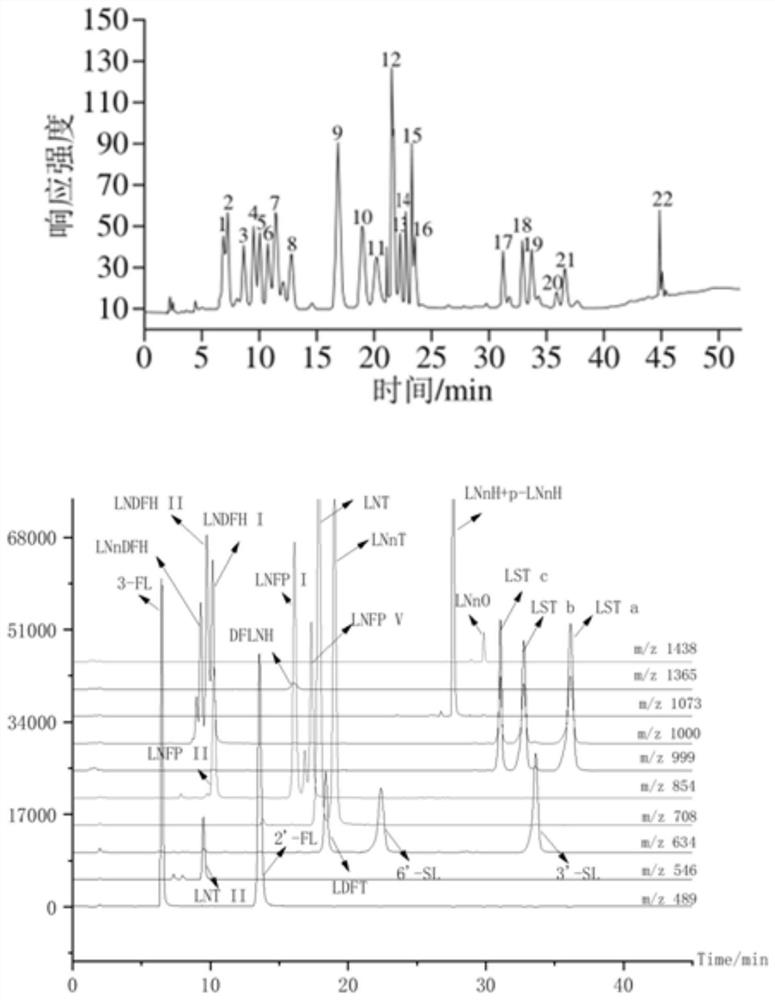
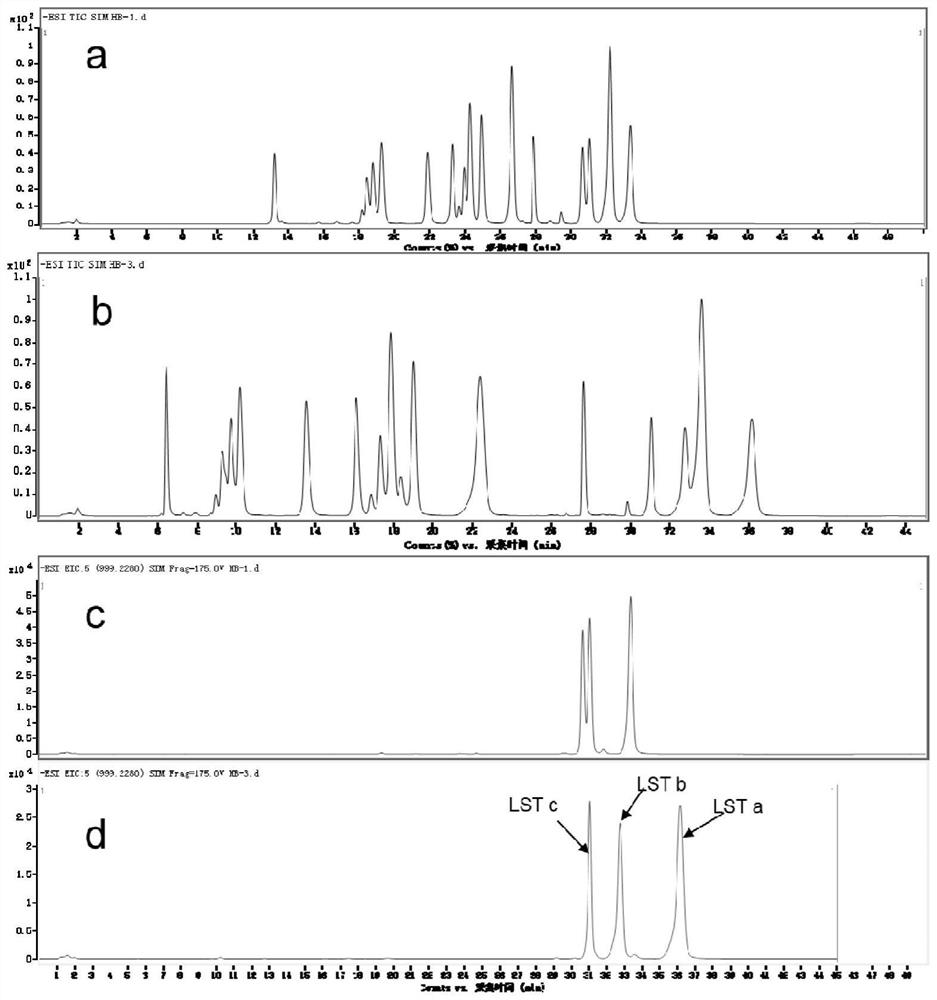
Method for detecting oligosaccharide in breast milk
A technology of breast milk oligosaccharide and detection method, applied in the detection field of breast milk oligosaccharide, can solve the problems of limited separation of oligosaccharide isomers and high limit of quantification, and achieves effective separation of various types and low limit of quantification , Pre-processing simple effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0085] Step 1. Take 200ul of breast milk sample, 13000r / min, centrifuge at 4°C for 15min, remove the fat in the upper layer, remove the supernatant, pass the supernatant through a 10kDa molecular weight filter, and centrifuge at 13000rpm / min for 30min; take 100μL of filtrate, add 100 μL of sodium borohydride (0.5M) was reacted in a water bath at 60°C for 30 minutes, 100 μL of acetic acid (0.5M) was added to stop the reaction, and the sample was filtered through a 0.22 μm membrane to obtain the sample.
[0086] Step 2. Take 1 mg of the purchased 21 kinds of oligosaccharide standard products, dissolve them in 2 mL of ultrapure water, and mix them to obtain a primary standard solution, which is stored at -80°C.
[0087] Step 3, take group 1 standard products including lactoyl-N-tetrasaccharide, lactoyl-N-neoctaose, 2'-fucose lactose, 3-fucose lactose, difucosyllactose, lactoyl -N-fucopentaose I, lactoyl-N-difucohexaose I, lactose-N-difucosyl-octasaccharide, 6'-sialylated lactose,...
Embodiment 2
[0093] Same as Example 1, the difference is only in the sample pretreatment, specifically:
[0094] Step 1. Take 300ul of breast milk sample, centrifuge at 13000r / min for 15min at 4°C, remove the fat in the upper layer, remove the supernatant, pass the supernatant through a 10kDa molecular weight filter, centrifuge at 13000rpm / min for 30min, take 150μL of filtrate, and then Add 150 μL of sodium borohydride (0.75M) to react in a 60°C water bath for 30 min, add 150 μL of acetic acid (0.75M) to stop the reaction, and filter through a 0.22 μm filter to obtain the sample.
[0095] Step 2. Take 1 mg of the purchased 21 kinds of oligosaccharide standard products, dissolve them in 2 mL of ultrapure water, and mix them to obtain a primary standard solution, which is stored at -80°C. .
[0096] Step 3, take group 1 standard products including lactoyl-N-tetrasaccharide, lactoyl-N-neoctaose, 2'-fucose lactose, 3-fucose lactose, difucosyllactose, lactoyl -N-fucopentaose I, lactoyl-N-difu...
Embodiment 3
[0102] Same as Example 1, the difference is only in the sample pretreatment, specifically:
[0103] Step 1. Take a breast milk sample of 400ul, 13000r / min, centrifuge at 4°C for 15min, remove the fat in the upper layer, remove the supernatant, pass the supernatant through a 10kDa molecular weight filter, centrifuge at 13000rpm / min for 30min, take 200μL of filtrate, add 200 μL of sodium borohydride (1.0 M) was reacted in a 60°C water bath for 30 min, 200 μL of acetic acid (1.0 M) was added to terminate the reaction, and the sample was filtered through a 0.22 μm filter membrane.
[0104] Step 2. Take 1 mg of the purchased 21 kinds of oligosaccharide standard products, dissolve them in 2 mL of ultrapure water, and mix them to obtain a primary standard solution, which is stored at -80°C. .
[0105] Step 3, take group 1 standard products including lactoyl-N-tetrasaccharide, lactoyl-N-neoctaose, 2'-fucose lactose, 3-fucose lactose, difucosyllactose, lactoyl -N-fucopentaose I, lact...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com