Method for detecting mutation of multiple gene loci by using high-throughput sequencing technology
A technology for technical detection and site mutation, which is applied in the field of multi-gene site mutation detection using high-throughput sequencing technology, which can solve problems such as difficulty in detecting low-frequency or rare mutations, systematic errors, etc., so as to reduce detection costs and errors. rate, the effect of ensuring accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
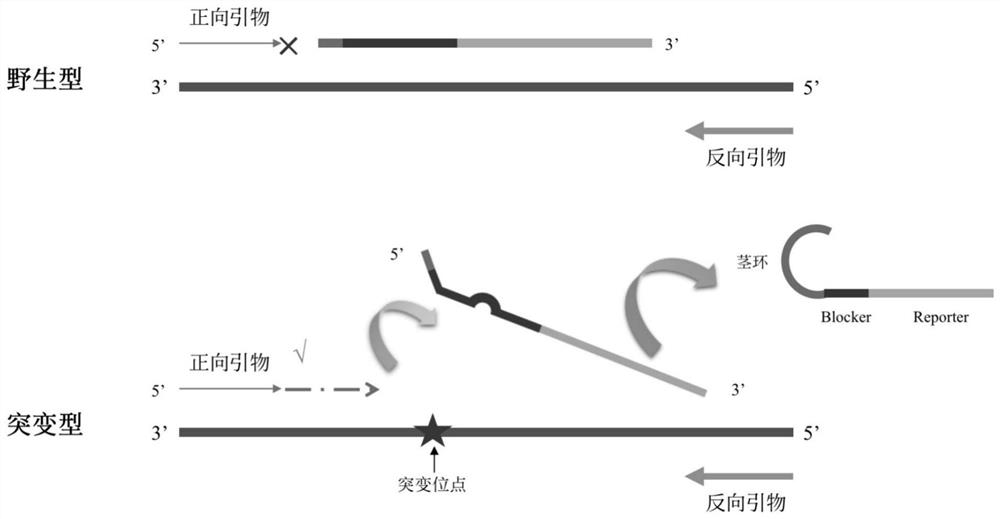
[0035] see figure 1 , this implementation discloses a specific probe structure, and the probe is sequentially Blocker and Reporter parts from the 5' end to the 3' end. The Tm value of the specific probe was 15-25°C higher than that of the universal primer at the I5 end.
[0036] The specific probe can be modified by any one of thio-modification, deoxyuracil modification, deoxyhypoxanthine modification, 2-methoxy modification and phosphorylation modification, or it can be modified in multiple ways at the same time.
[0037] Wherein, the length of the Blocker part is 15-20 bases, including artificially introduced 2-6 arbitrary base sequences, other sites are complementary to the wild-type DNA sequence to be tested, and the 5' end of the Blocker part introduces a stem-loop structure, And the length of the stem-loop structure is 3-10 bases, and its sequence is complementary to a part of the Blocker sequence; the Reporter part is 20-30 bases in length, and its sequence is compleme...
Embodiment 2
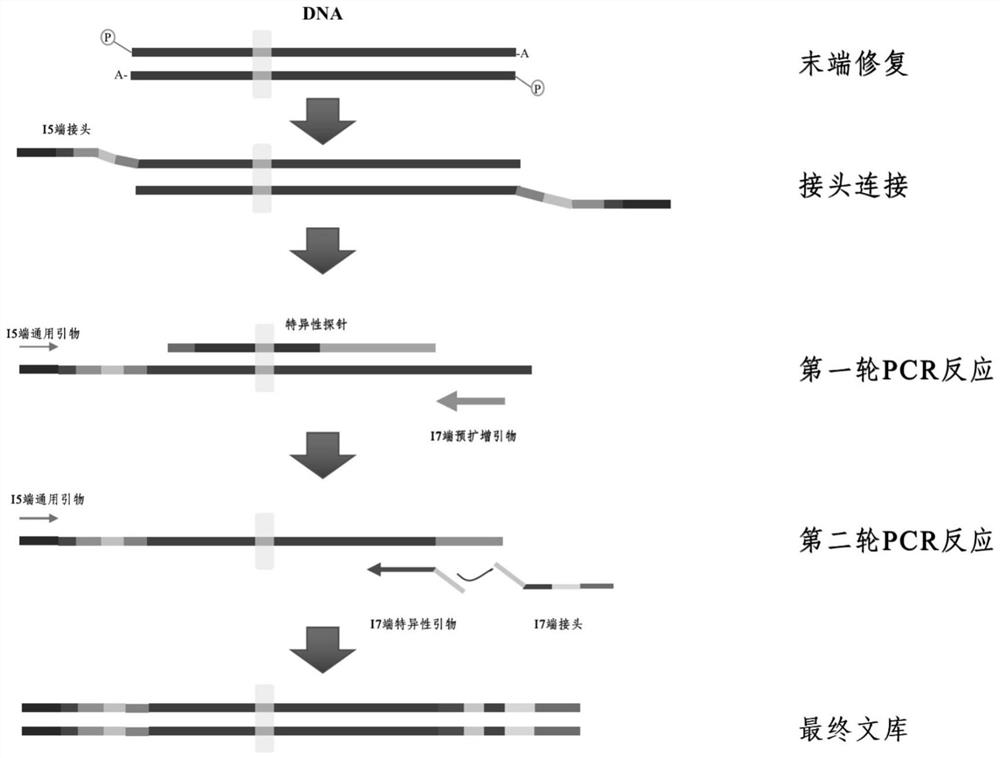
[0052] This embodiment specifically discloses a specific detection method by taking the detection of DNA mutation in peripheral blood circulating tumor cell samples as an example
[0053] 1) End repair
[0054] This part is divided into two steps. First, end-repair of cfDNA is performed under the action of DNA polymerase to make double-stranded DNA form blunt ends, and then phosphorylation at the 5' end and addition of dA tail at the 3' end are performed respectively.
[0055] Prepare the reaction system in the following table:
[0056] component volume cfDNA XμL Nuclease-free water (40-X)μL Fragment and End Repair Buffers 10μL Fragments and end repair enzymes 10μL total capacity 60μL
[0057] After the preparation is completed, mix well and centrifuge, and use a common PCR machine. The reaction procedure is shown in the following table:
[0058] temperature time 20℃ 30 minutes 65℃ 30 minutes 4℃ ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com