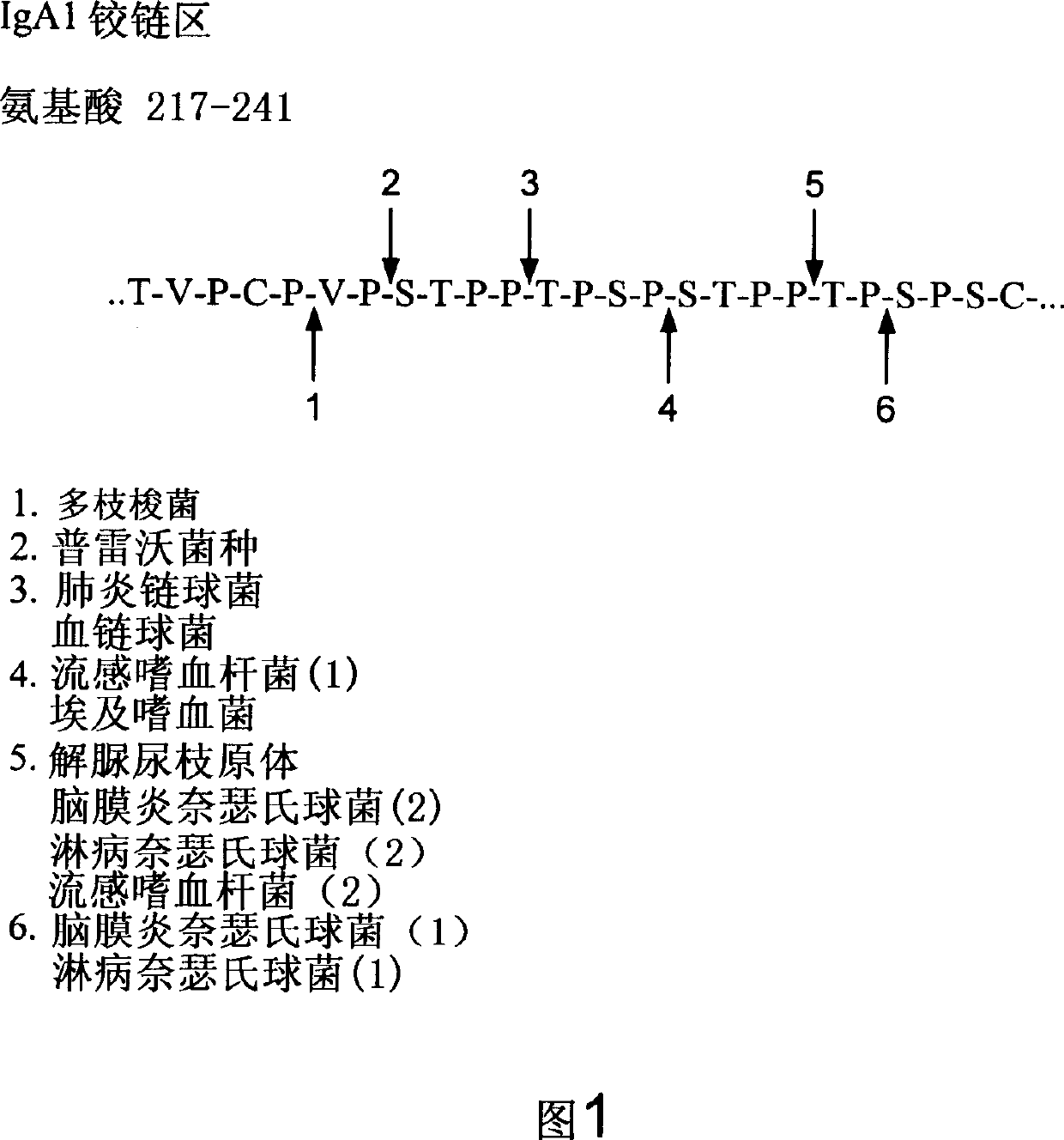
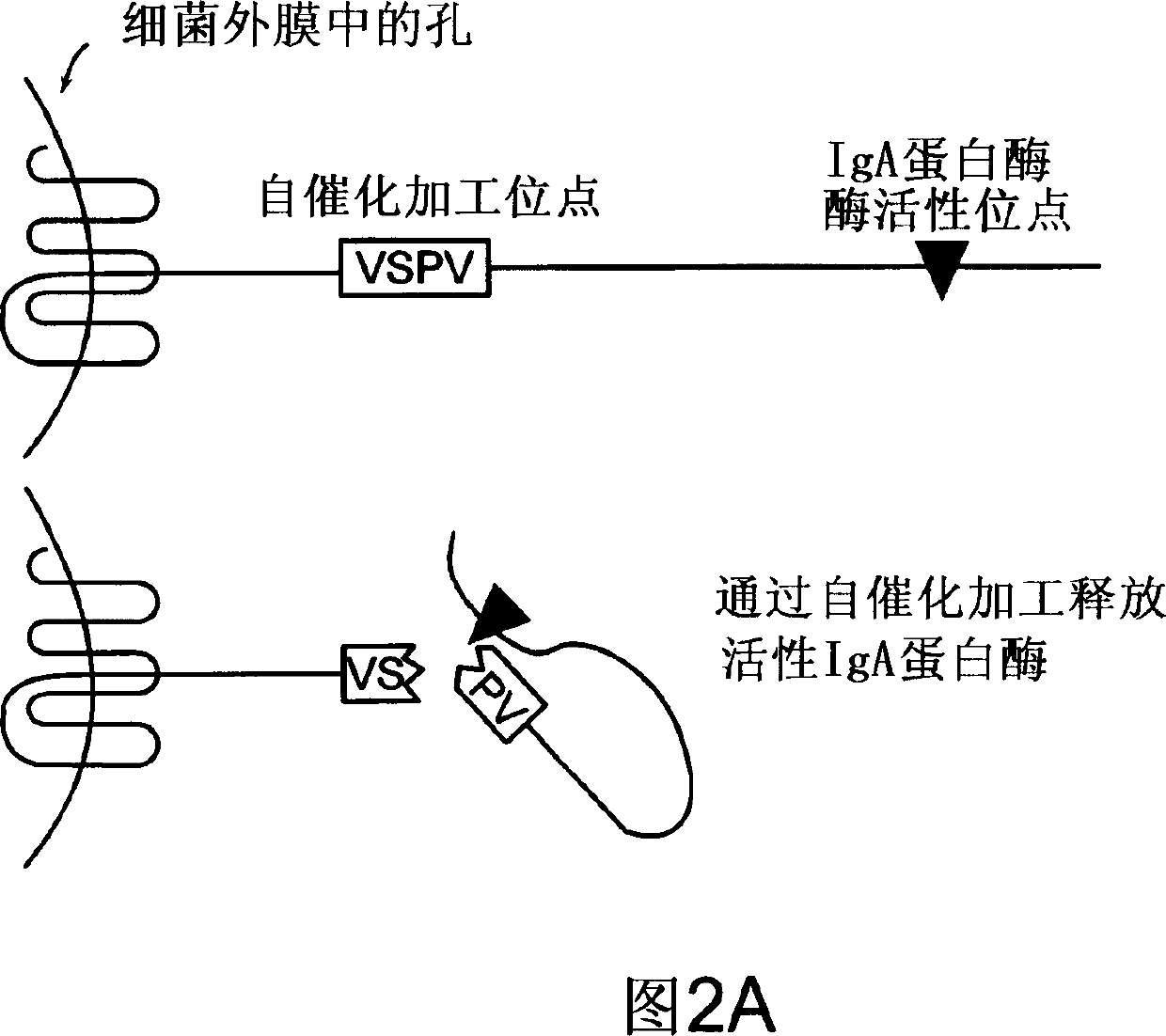
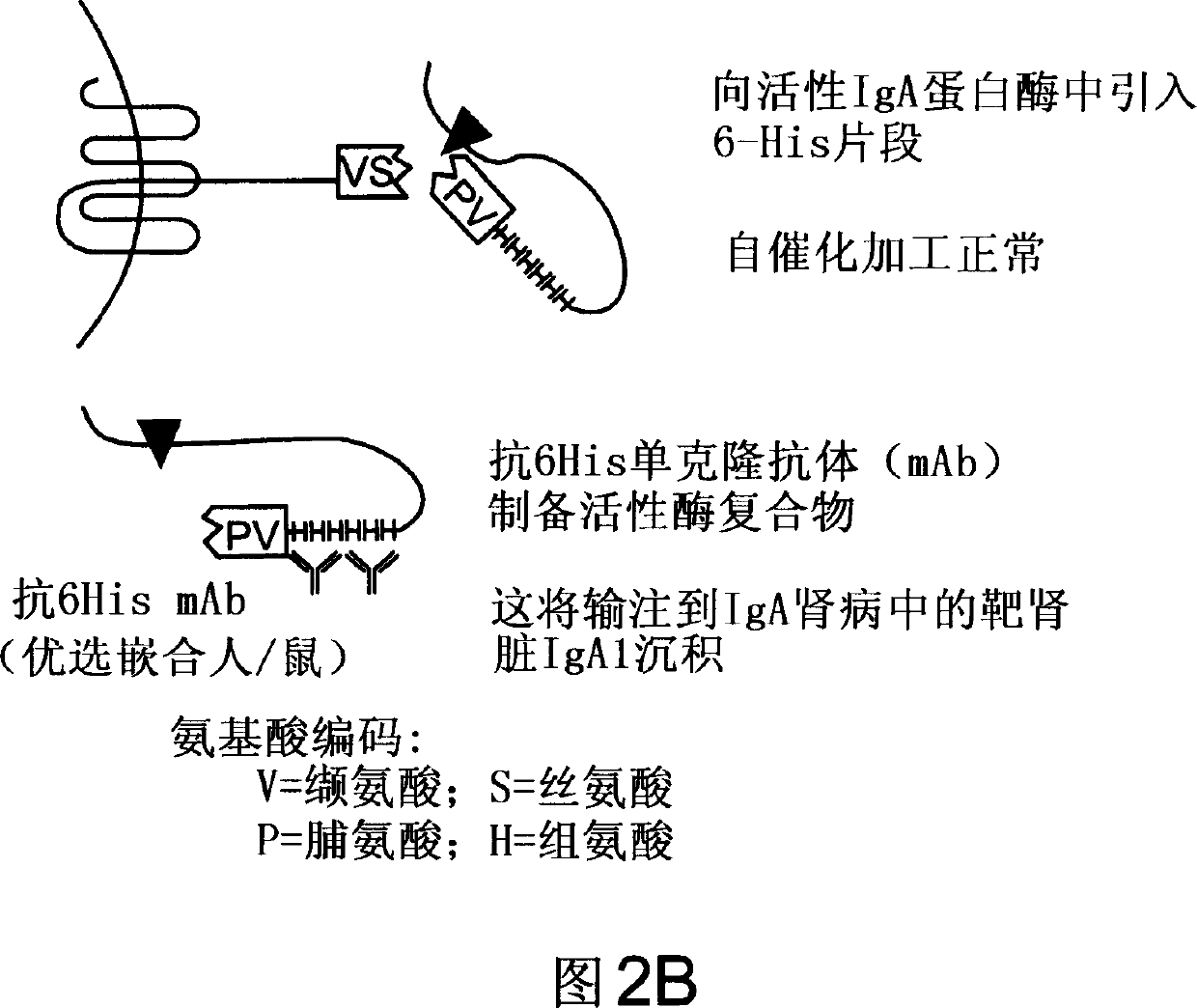
Treatment of IgA1 deposition diseases
A DNA sequence and coding technology, applied in skin diseases, blood diseases, bone diseases, etc., can solve problems such as non-specific IgA1 molecules
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0143] Example 1. Construction of tagged IgA1 protease
[0144] The His-tag was fused in frame to the H. influenzae IgA1 protease by PCR-based site-directed mutagenesis using plasmid pFG26 containing the DNA sequence encoding the H. influenzae IgA1 protease. As shown in Figure 4, two PCR fragments were generated from pFG26. Using the oligonucleotide primers "HFD6His1" (Primer 1) and "HFD6His2" (Primer 2) shown below generated the first fragment, the XbA1 and pml1 fragments. A second fragment, the pmL I and Acc I fragments, was generated using primers 3 and 4 shown below.
[0145] Primer 2: HFD-5XbaI: 5'GATCCGCTTACCAATTATGC 3' (SEQ ID NO: 20)
[0146] Primer 2: HFD6His1:
[0147] 5'CTTGGTACGCTAGGCACGTGATGATGATGATGATGAGGTGTTGTGATATTT
[0148] GTCG-3' (SEQ ID NO: 21)
[0149] Primer 2: HFD6His2: 5'-CCTAATAATATTCAAGCTCACGTGCCTAGCGTACC-3'
[0150] (SEQ ID NO: 22)
[0151] Primer 2: HFD-F-ACCI: 5'-TTCAGCAGAAGTCTCTTGC-3' (SEQ ID NO: 23)
[0152]After amplifying the two fragme...
Embodiment 2
[0153] Example 2. Generation of bacterial strains expressing tagged IgA1 protease
[0154] Haemophilus influenzae bacterial strains expressing only the tagged, enzymatically active IgA1 protease were generated by conventional recombinant techniques. Briefly, the plasmid pJQ / Rd6His produced in Example 1 was cut with restriction enzymes Cla I and NdeI. The gene was isolated and transformed into the Haemophilus influenzae Rd strain (Rd3-13) producing an enzymatically inactive IgA1 protease (Plaut AG, Qiu, J, Grundy, F. and Wright, A.J Infect Dis. (1992) Jul. ; 166(1):43-52), so that the His-tagged IgAl protease is inserted into the bacterial genome by recombination. Bacteria for restoring enzymatic activity were screened by verifying the presence of active protease in the bacterial growth medium of selected clones using human IgAl as a substrate.
[0155] Introduction of the 6-His mutation into the active enzyme was confirmed using PCR fragments of genomic DNA to verify the pre...
Embodiment 3
[0159] The therapeutic effect of IgA1 protease for treating IgA nephropathy can be verified in the IgA nephropathy mouse model.
[0160] Multiple Swiss-Webster mice (Charles River Laboratories) were used per experiment and grouped (typically 10 mice / group). The mice were first placed in metabolic cages and urine was collected for 24 hours to measure the amount of hematuria (red blood cells in the urine) and proteinuria (proteins in the urine) in the urine. This provides baseline values for hematuria and proteinuria in normal healthy mice. IgA nephropathy was then induced in mice as described in Gesualdo L. et al, (1990) J. Clin. Invest. 86:715-722.
[0161] After inducing nephropathy, mice were intraperitoneally injected with multiple doses of saline (control), inactive IgA1 protease (control), active IgA1 protease, and IgA1 protease complexed with immunoglobulin (such as complexed with anti-tag antibody or anti-IgA1 protease antibody). An exemplary dosing regimen includes...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com