High efficiency full genome fixing method
A whole genome, fixed method technology, applied in the field of genome detection in biomedicine, can solve the problems of limited detection sites and incomplete information, and achieve the effects of saving detection costs, accurate information, and saving preparation time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0033] Example 1 Yeast whole genome and preparation method of whole genome DNA chip
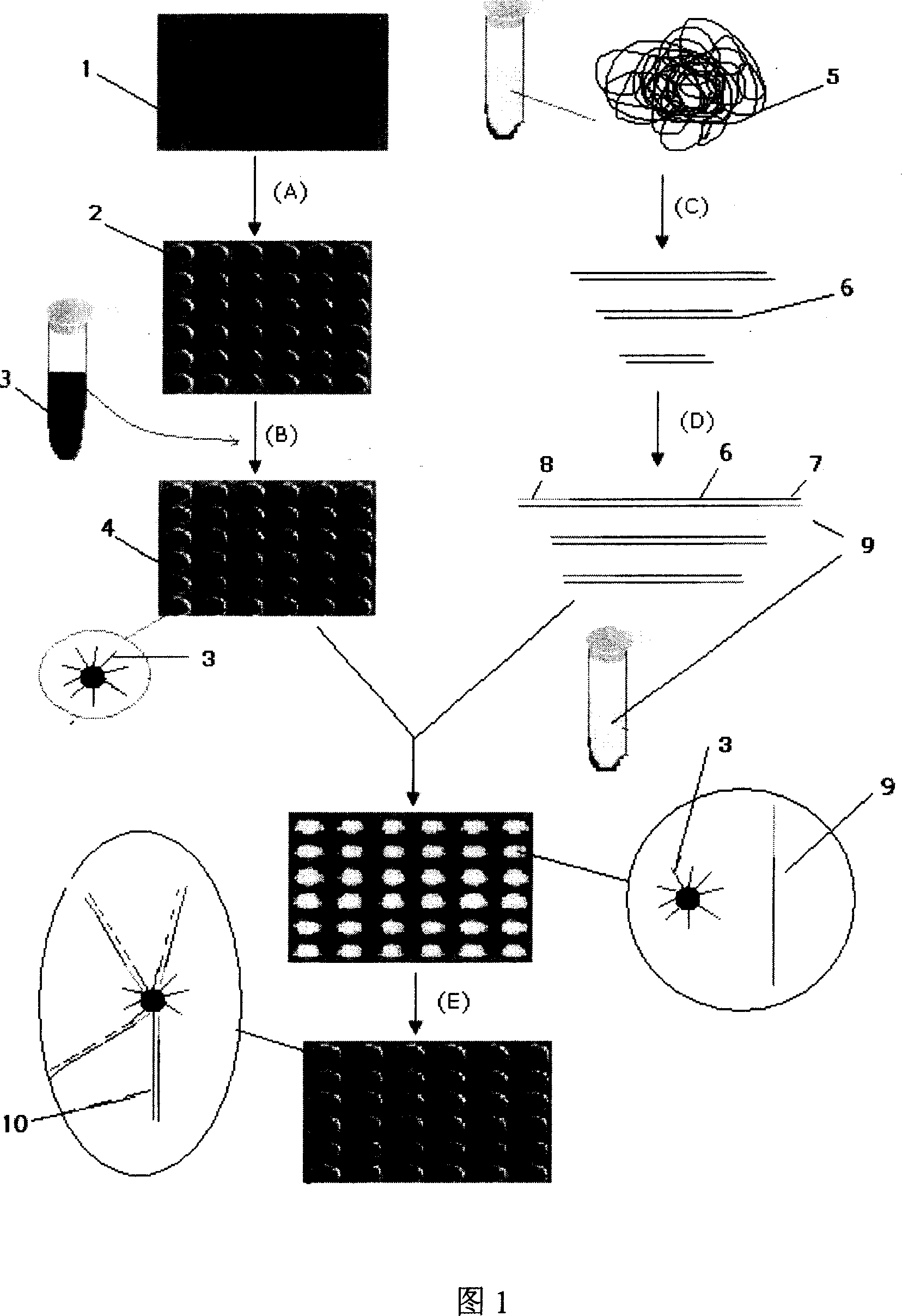
[0034] Referring to accompanying drawing 1, on a flat glass slide, process by photolithography into the microporous plate of diameter 50 microns, depth 50 microns, then use dichlorodimethylsilane to carry out hydrophobic treatment on the surface of microporous plate. After the micropores were washed with water and dried, they were modified with 5% 3-aminopropyltrimethoxysilaneacetone for 1 hour, and treated with 5% glutaraldehyde for 2 hours, and then reacted with the oligonucleotide solution modified with the amino group at the 5' end. The oligonucleotides are immobilized in the microwells. On the other hand, the yeast genome is cut into fragments with a size of about 500 bases by enzymes, and these fragmented nucleic acid sequences are connected with a pair of universal linkers under the action of ligase, one of which is connected to the microwell The oligonucleotide molecular sequence imm...
example 2
[0037] Example 2 The preparation method of human whole genome and its whole genome DNA chip
[0038] Referring to accompanying drawing 1, a microporous plate with a diameter of 50 microns and a depth of 50 microns is processed by photolithography on a flat glass sheet, and then the surface of the microporous plate is subjected to hydrophobic treatment with dichlorodimethylsilane. Mix oligonucleotides modified with acrylamide groups at the 5' end with acrylamide monomers, ammonium persulfate, etc., and fill them in micropores, and let the water in the prepolymer volatilize naturally. When the water volatilizes to the prepolymer When only two-thirds to one-third of the micropores are occupied, the glass sheet is placed under vacuum, and the polymerization reaction is promoted to form a gel under the vaporized atmosphere of the initiator and fixed in the micropores.
[0039] On the one hand, the human genome is cut into fragments with a size of 50-1000 bases with enzymes, and the...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| depth | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com