Methods and compositions for analysis of plant gene function
a technology of plant gene function and composition, applied in the field of molecular biology, can solve the problems of large amount of time required to obtain, and no virus is currently available as a vector,
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Identification and Isolation of F-BMV
[0051] An uncharacterized virus was isolated from an infected tall fescue plant obtained from a breeder's stock of fescue lines at the University of Missouri and maintained in a greenhouse. The virus was determined to be an isolate of BMV by electron microscopy, western blot assay using an antibody against BMV coat protein and northern blot assay using an RNA profile specific for the conserved 3′ untranslated region of the BMV genomic RNAs. Because this virus infected tall fescue, it was designated F-BMV. R-BMV was from a previously described source and maintained in a growth chamber at the Noble Foundation (Ding et al., 1999).
[0052] An initial study was made of the host range of the virus. Plants of three rice cultivars were inoculated with F-BMV or R-BMV virion and grown inside a greenhouse at 25° C. The inoculated plants were observed for disease symptoms for 28 days and tested for virus infection by immunocapture RT-PCR using an antibody ag...
example 2
Cloning and Sequencing of F-BMV and R-BMV
[0053] Virions of F-BMV and R-BMV (Ding et al., 1999) were purified from systemically infected barley leaves by PEG precipitation and differential centrifugation (Lane, 1981). Viral RNA was then isolated from purified virions through phenol / chloroform extraction and ethanol precipitation. First-strand cDNA of RNAs 1, 2 and 3 representing each virus genome were synthesized by priming viral RNAs with Primer HK-R (5′-GACAATGGTCTCTTTTAGAG-3′) (SEQ ID NO:5). The ten 5′-most nucleotides of the HK-R primer sequence contain a PshA1 restriction site coincident with the 3′ end of the R-BMV RNA sequence. Full-length PCR products of the RNAs 1, 2 and 3 were synthesized for each virus using the HK-R primer and primers containing sequences corresponding to the T3 promoter sequence and the 5′ end of the respective R-BMV RNA sequences: (i.e., HK-1F, 5′-AATTAACCCTCACTAAAGGGAGAGTAGACCACGGAACGAGGT-3′ for RNA 1 (SEQ ID NO:6); HK-2F, 5′-AATTAACCCTCACTAAAGGGAGAGT...
example 3
Production of Infectious Transcripts from cDNAs of F-BMV and R-BMV
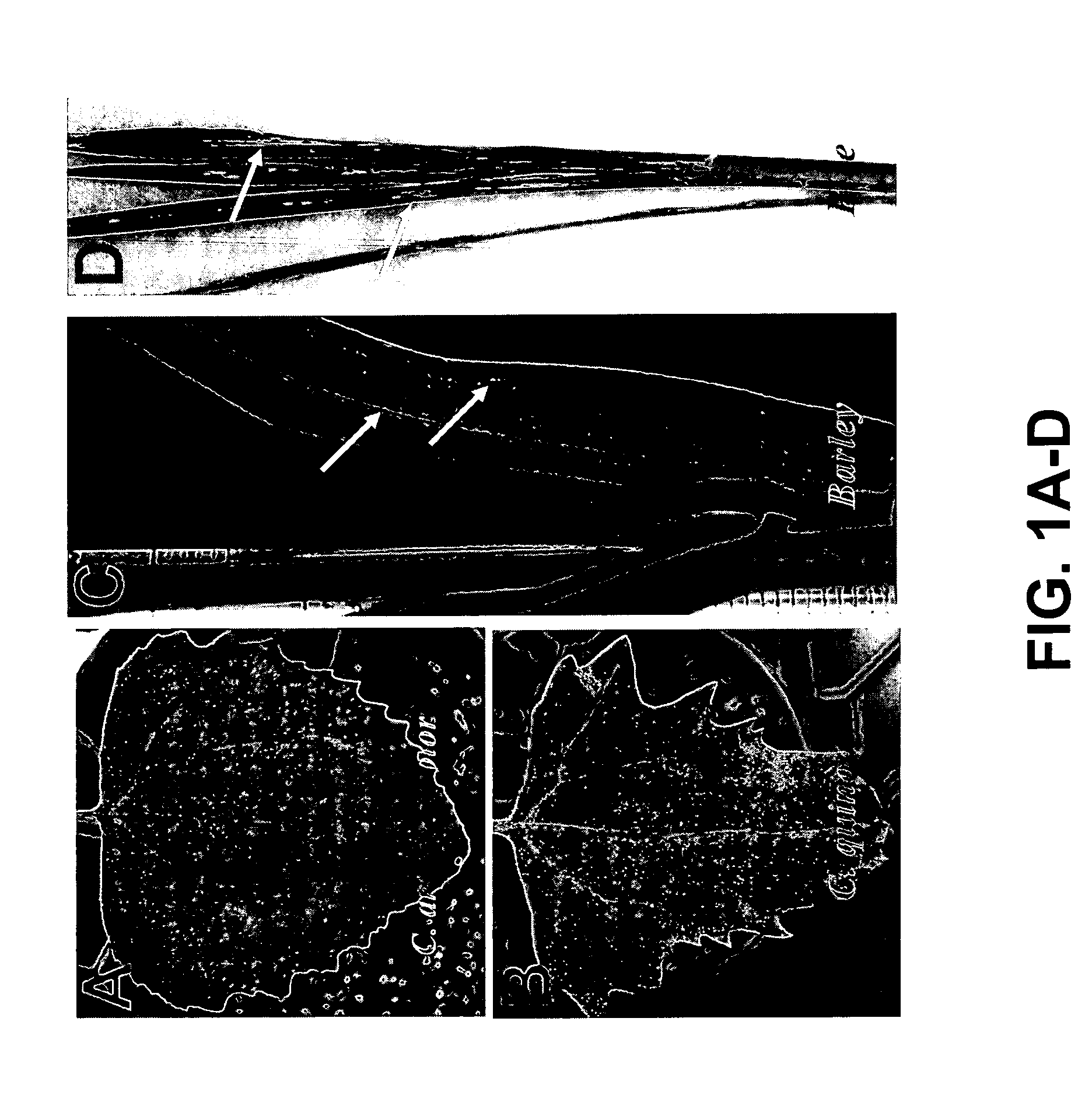
[0055] Plasmid DNAs representing RNA 1 of F-BMV (pF1-11) and R-BMV (pR1-26) were linearized with the restriction enzyme SpeI. Plasmid DNAs representing RNA 2 and 3 of the F-BMV (pF2-2 and pF3-5) and R-BMV (pR2-9 and pR3-3) were linearized with the restriction enzyme PshA. In vitro transcripts were synthesized from each clone using the mMeSSAGE mMACHINE transcription kit as described (Ambion, Austin, Tex.). The three RNA transcripts representing their respective viruses (i.e., F-BMV or R-BMV) were equally mixed and inoculated to leaves of Hordeum vulgare (barley) cv. Morex, Chenopodium amaranticolor and C. quinoa (5 to 6 μl mixed RNA transcripts / leaf, one leaf / plant). The inoculated plants were grown inside a growth chamber at 22 / 18° C. (day / night). At 3 to 4 days post inoculation (dpi), C. amaranticolor and C. quinoa leaves showed chlorotic lesions (FIG. 1A-1B). Barley plants inoculated with the transcripts represent...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com