Small molecule printing
a technology of small molecules and printing, applied in the field of small molecules printing, can solve the problems of slow serial process and limited to only a few iterations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Small Molecule Printing using Michael Addition
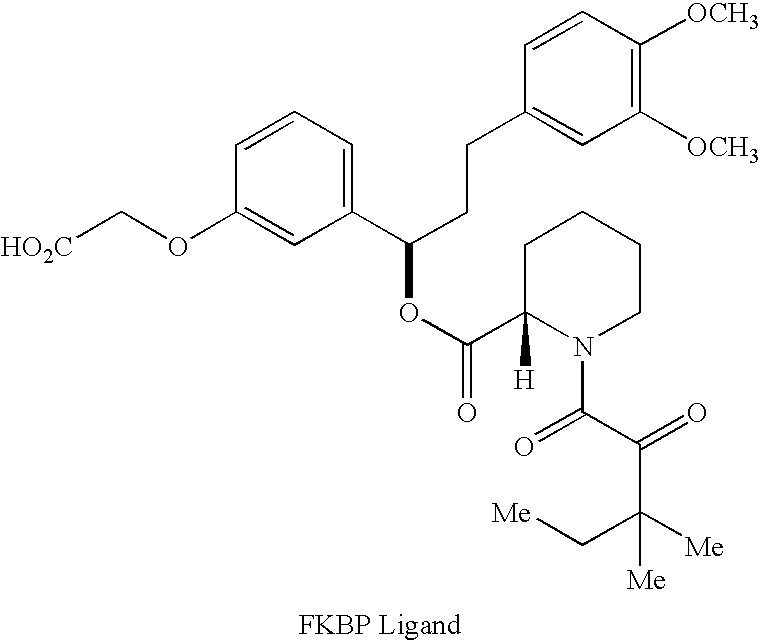
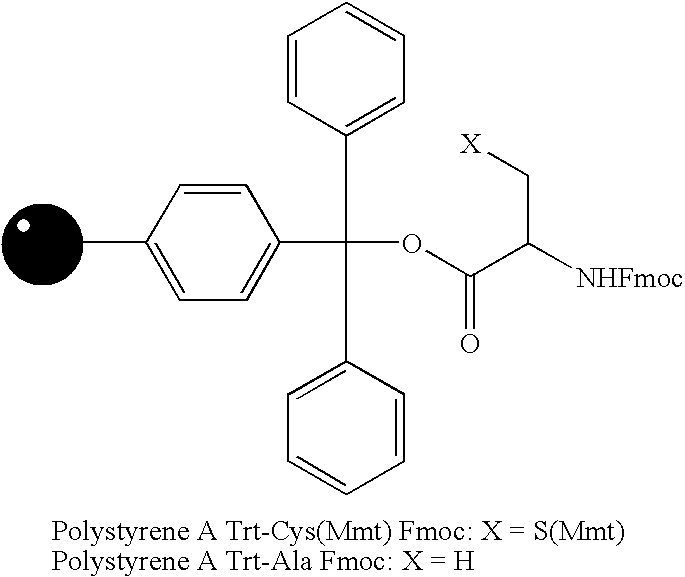
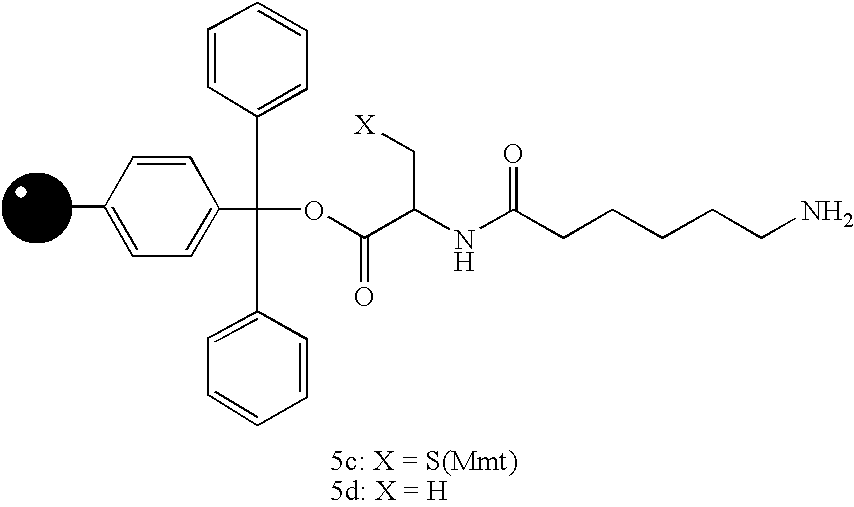
[0054] In order to demonstrate the utility of small molecule printing as a technique identifying small molecule-protein interactions, three unrelated molecules were chosen for which specific protein receptors are available. Compound 1 (FIG. 6, R═OH) is the vitamin biotin, which is recognized by the bacterial protein streptavidin (Chaiet et al., Arch. Biochem. Biophys. 1964, 106, 1; incorporated herein by reference). Compound 2 (R═OH) is a derivative of the steroid digoxigenin and is recognized by the mouse monoclonal antibody DI-22 (Sigma). Finally, compound 3 (R═OH) is a synthetic pipecolyl α-ketoamide, which was designed to be recognized by the human immunophilin FKBP12 (Holt et al., J. Am. Chem. Soc. 1993, 115, 9925; incorporated herein by reference). Each of these compounds was attached to 400-450 μm diameter polystyrene beads (estimated capacity of 20 nmol per bead) via a 6-aminocaproic acid linker and either 4-methoxytrityl-protec...
example 2
Small Molecule Printing Using Silylation Reaction
[0058] Standard glass slides were activated for selective reaction with alcohols (FIG. 9). Microscopic slides were first treated with a H2SO4 / H2O2 solution (“piranha”) for 16 hours at room temperature. After extensive washing with water, the slides were treated with thionyl chloride and a catalytic amount of DMF in THF for 4 hours at room temperature. Surface characterization by x-ray photoelectron spectroscopy (XPS) confirmed the presence of chlorine on the slide (Strother et al., J. Am. Chem. Soc., 2000, 122, 1205-1209; incorporated herein by reference). To test the ability of these chlorinated slides to capture alcohols released from synthesis beads, we initially used three alcohol-containing small molecules and a bead linker reagent developed for chemical genetic applications of diversity-oriented synthesis.
[0059] Primary alcohol derivatives of a synthetic α-ketoamide (Holt et al., J. Am. Chem. Soc. 1993, 115, 9925-9938; incorpo...
example 3
Fabrication of Custom Slide Reaction Vessels
[0062] In an effort to minimize reagent volume during the chemical treatment of glass microscope slides, we designed and fabricated custom slide-sized reaction vessels that enable the uniform application of ˜1.4 mL solution to one face of a 2.5 cm×7.5 cm slide. First, a master template mold was cut from a block of Delhran plastic according to the blueprint shown in FIG. 13. The slide-sized reaction vessels were prepared by casting degassed polydimethysiloxane (PDMS, Sylgard Kit 184, Dow corning, Midland, Mich.) prepolymer around the master template in a polystyrene OmniTray (Nalge Nunc International, Naperville, Ill.). After curing for four hours at 65° C., the polymer was peeled away from the master to give the finished product (FIG. 14).
[0063] To use the vessels, slides were placed face-down as illustrated below and reagent was injected under the slides with a P1000 Pipetman (FIG. 15).
PUM
| Property | Measurement | Unit |
|---|---|---|
| distance | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
| volume | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com