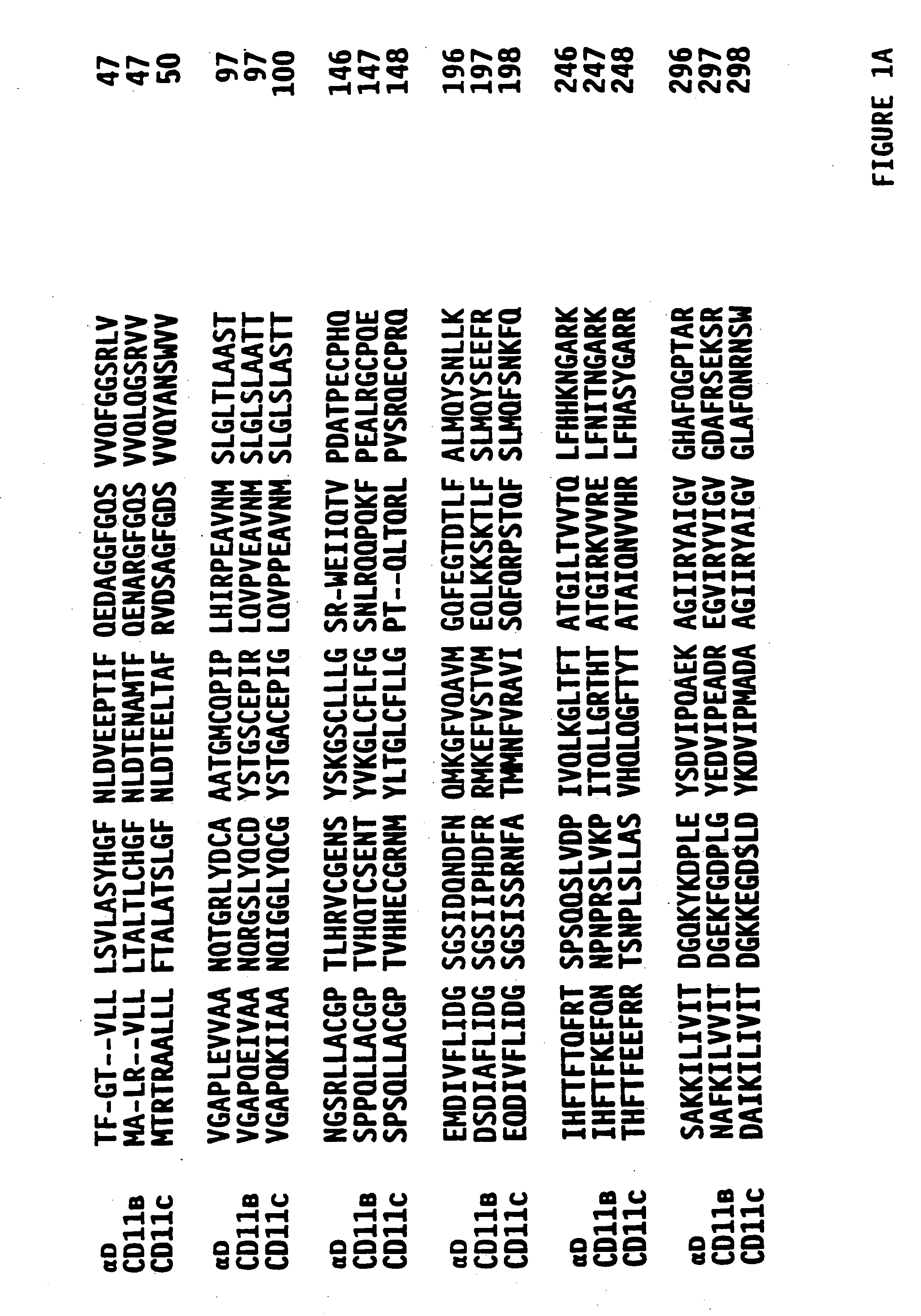
Novel human beta-2 integrin alpha subunit
a human beta2 and alpha subunit technology, applied in the field of new human beta2 integrin alpha subunits, can solve the problem of inability to define the correlation between human beta2 integrin subunits and those identified in other species
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Attempt to Detect a Human Homolog of Canine αTM1
[0055] The monoclonal antibody Ca11.8H2 [Moore, et al., supra] specific for canine αTM1 was tested for cross-reactivity on human peripheral blood leukocytes in an attempt to identify a human homolog of canine αTM1. Cell preparations (typically 1×106 cells) were incubated with undiluted hybridoma supernatant or a purified mouse IgG-negative control antibody (10 μg / ml) on ice in the presence of 0.1% sodium azide. Monoclonal antibody binding was detected by subsequent incubation with FITC-conjugated horse anti-mouse IgG (Vector Laboratories, Burlingame, Calif.) at 6 μg / ml. Stained cells were fixed with 2% w / v paraformaldehyde in phosphate buffered saline (PBS) and were analyzed with a Facstar Plus fluorescence-activated cell sorter (Becton Dickinson, Mountain View, Calif.). Typically, 10,000 cells were analyzed using logarithmic amplification for fluorescence intensity.
[0056] The results indicated that Ca11.8H2 did not cross-react with ...
example 2
Affinity Purification of Canine αTM1 for N-Terminal Sequencing
[0058] Canine αTM1 was affinity purified in order to determine N-terminal amino acid sequences for oligonucleotide probe / primer design. Briefly, anti-αTM1 monoclonal antibody Ca11.8H2 was coupled to Affigel® 10 chromatographic resin (BioRad, Hercules, Calif.) and protein was isolated by specific antibody-protein interaction. Antibody was conjugated to the resin, according to the BioRad suggested protocol, at a concentration of approximately 5 mg antibody per ml of resin. Following the conjugation reaction, excess antibody was removed and the resin blocked with three volumes of 0.1 M ethanolamine. The resin was then washed with thirty column volumes of phosphate buffered saline (PBS).
[0059] Twenty-five grams of a single dog spleen were homogenized in 250 ml of buffer containing 0.32 M sucrose in 25 mM Tris-HCl, Ph 8.0, with protease inhibitors. Nuclei and cellular debris were pelleted with centrifugation at 1000 g for 15...
example 3
Large Scale Affinity Purification of Canine αTM1 for Internal Sequencing
[0069] In order to provide additional amino acid sequence for primer design, canine αTM1 was purified for internal sequencing. Three sections of frozen spleen (approximately 50 g each) and frozen cells from two partial spleens from adult dogs were used to generate protein for internal sequencing. Fifty grams of spleen were homogenized in 200-300 ml borate buffer with a Waring blender. The homogenized material was diluted with 1 volume of buffer containing 4% NP-40, and the mixture then gently agitated for at least one hour. The resulting lysate was cleared of large debris by centrifugation at 2000 g for 20 min, and then filtered through either a Corning (Corning, N.Y.) prefilter or a Corning 0.8 micron filter. The lysate was further clarified by filtration through the Corning 0.4 micron filter system.
[0070] Splenic lysate and the antibody-conjugated Affigel® 10 resin described in Example 2 were combined at a 1...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com