Bacterial signaling molecules that down-regulate pathogenic bacterial virulence properties
a signaling molecule and pathogenic bacteria technology, applied in the field of bacteria proteins, peptides and amino acids, can solve the problems of poor penetration at the tissue or biomaterial interface, inability to eradicate, and ineffective agents, and achieve the effect of increasing the proliferation ra
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
[0059] Virulence factors (VFs) produced by bacteria are exemplified below, in which the expression of virulence factors in a number of UPEC strains was confirmed (see Table 1). Oligonucleotide primers specific for the genes encoding VFs, such as type 1, P, F1 C and S fimbriae, haemolysin A, aerobactin, afimbrial adhesin I (afa I), cytotoxic necrotizing factors I and II (cnfs I and II), KII and KIII capsular proteins and 16S rRNA (control), were generated and PCR was carried out to screen for the presence of these genes. One PCR product for each VF was sequence verified. To confirm the PCR results, sequenced amplicons were also DIG-labelled and used as probes to screen genomic DNA of all the E. coli strains by dot-blot hybridization. The expression of haemolysin A and type 1 pili were also determined by plating on Columbia agar and haemagglutination assays, respectively.
TABLE 1Bacterial Strains Used in This StudyPreviouslyIdentifiedBacterial SpeciesStrainHuman OriginVirulence Facto...
example 2
[0061] To initially assess the effects of Lactobacillus secreted by-products on UPEC growth, differential antagonism and well diffusion qualitative assays were employed. The Lactobacillus strains Lactobacillus rhamnosus GR-1 and L. reuteri RC-14 were grown as 1 cm wide lawns on BHIS agar for 48 hours. These lawns were then removed, perpendicular 1 cm lawns of each UPEC strain were plated across the original Lactobacillus streaks and plates were incubated 16-18 hours and the growth was assessed. Well diffusion assays involved testing spent culture supernatants (SCS) isolated from both Lactobacillus strains grown for 48 hours in BHIS broth on the growth of the UPEC strains. Lactobacillus SCS were pipetted into wells cut out of agar plates harboring surface lawns of the UPEC strains. The plates were incubated for 16-18 hours and the growth was assessed. Additionally, supernatants were either boiled for 10 minutes, neutralized to pH 7.0, treated with catalase or treated with proteinase ...
example 3
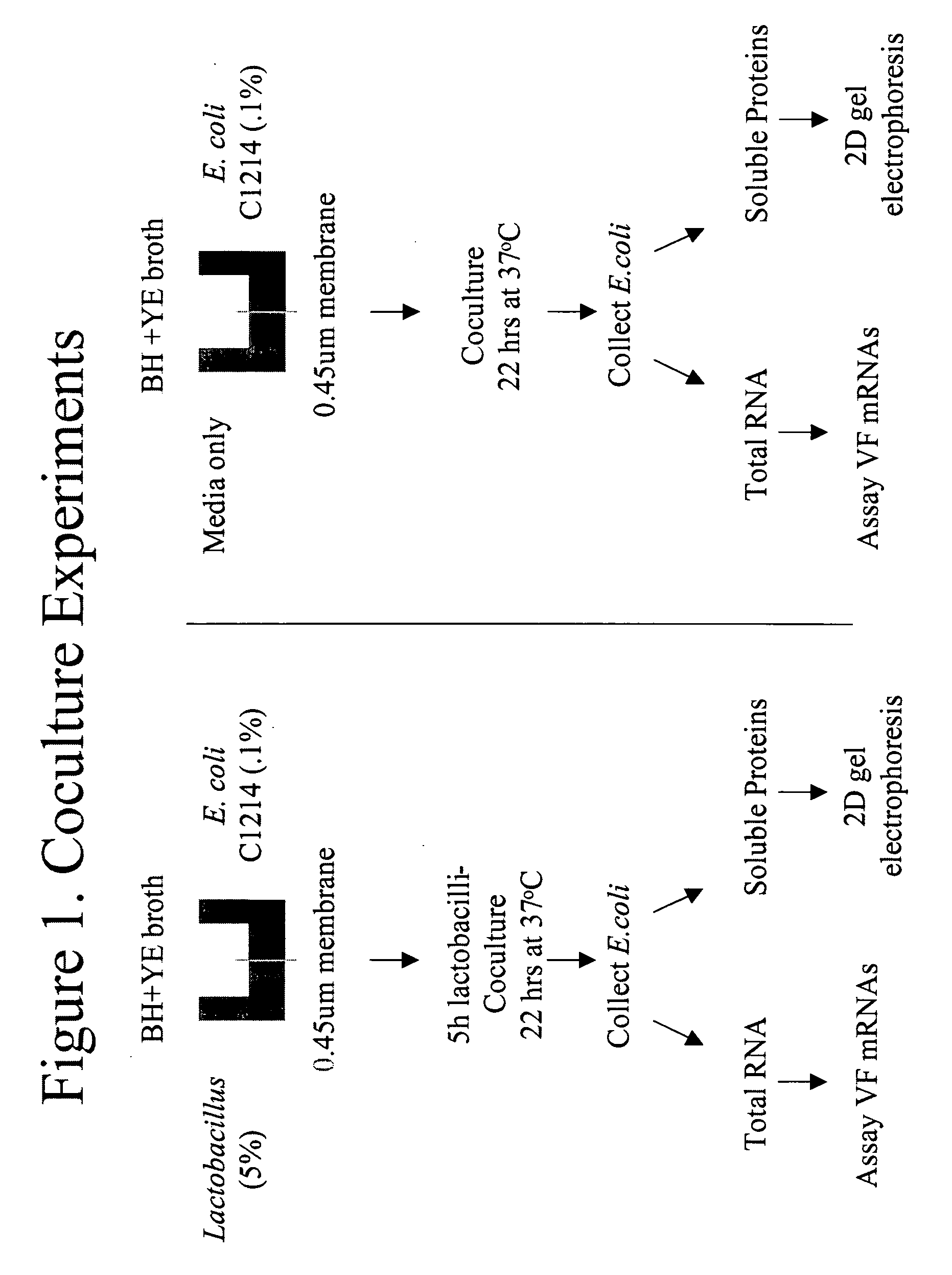
[0065] Using an experimental set up shown in FIG. 1, the ability of Lactobacillus-derived substances to affect the virulence of E. coli was demonstrated.
[0066] As shown in FIG. 1, the coculture of Lactobacillus and E. coli are separated by a 0.45 um membrane, which only allows molecules, not cells, to pass through. Thus, only by-products of the bacteria were able to induce the reaction or the bacterial “cross talk.” The first evidence of such bacterial “cross-talk” was obtained from a dot blot, which showed changes in S fimbrial expression caused by incubation with Lactobacillus GR-1. Similar changes were also observed from SQ-RT-PCR results, which showed that expression of VFs were altered, either up regulated or down regulated, when the E. coli strains were co-cultured with Lactobacillus GR-1 or RC-14. FIG. 4 to FIG. 9 illustrate the 2D protein gel results, which demonstrated that the expression of a uropathogenic E. coli VFs, such as fimbria, were altered when the E. coli were c...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| time period | aaaaa | aaaaa |
| antibiotic-resistance | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com