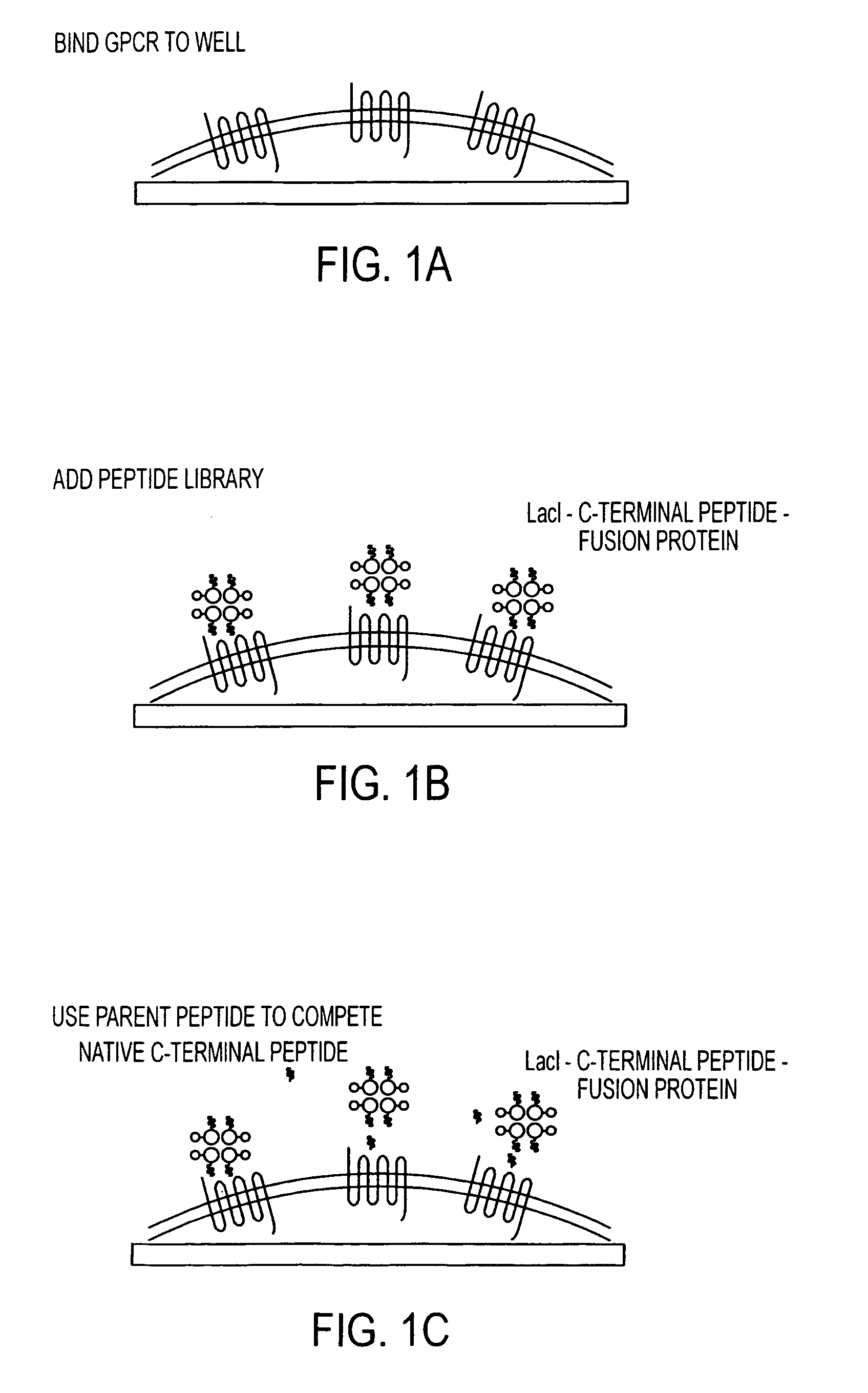
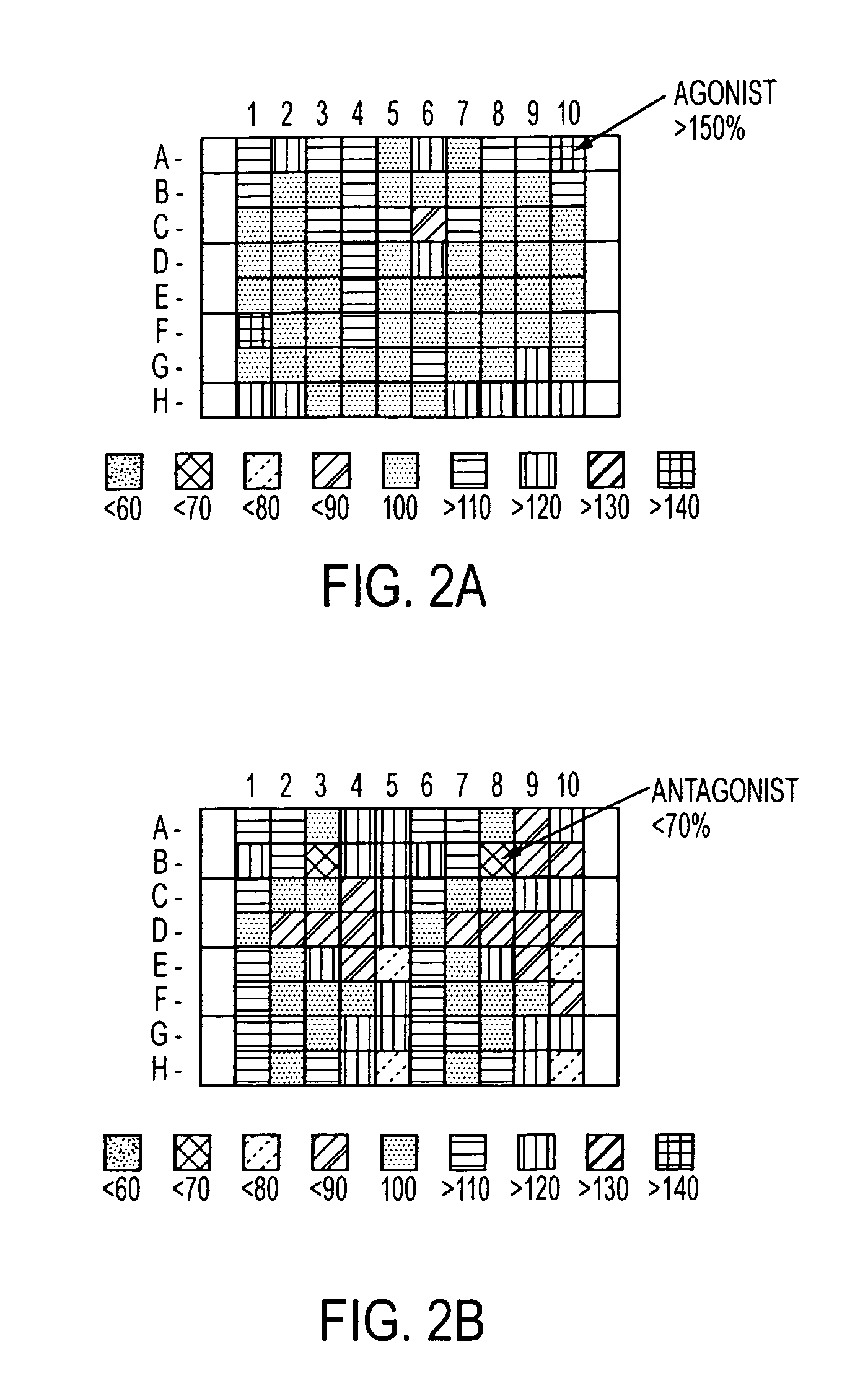
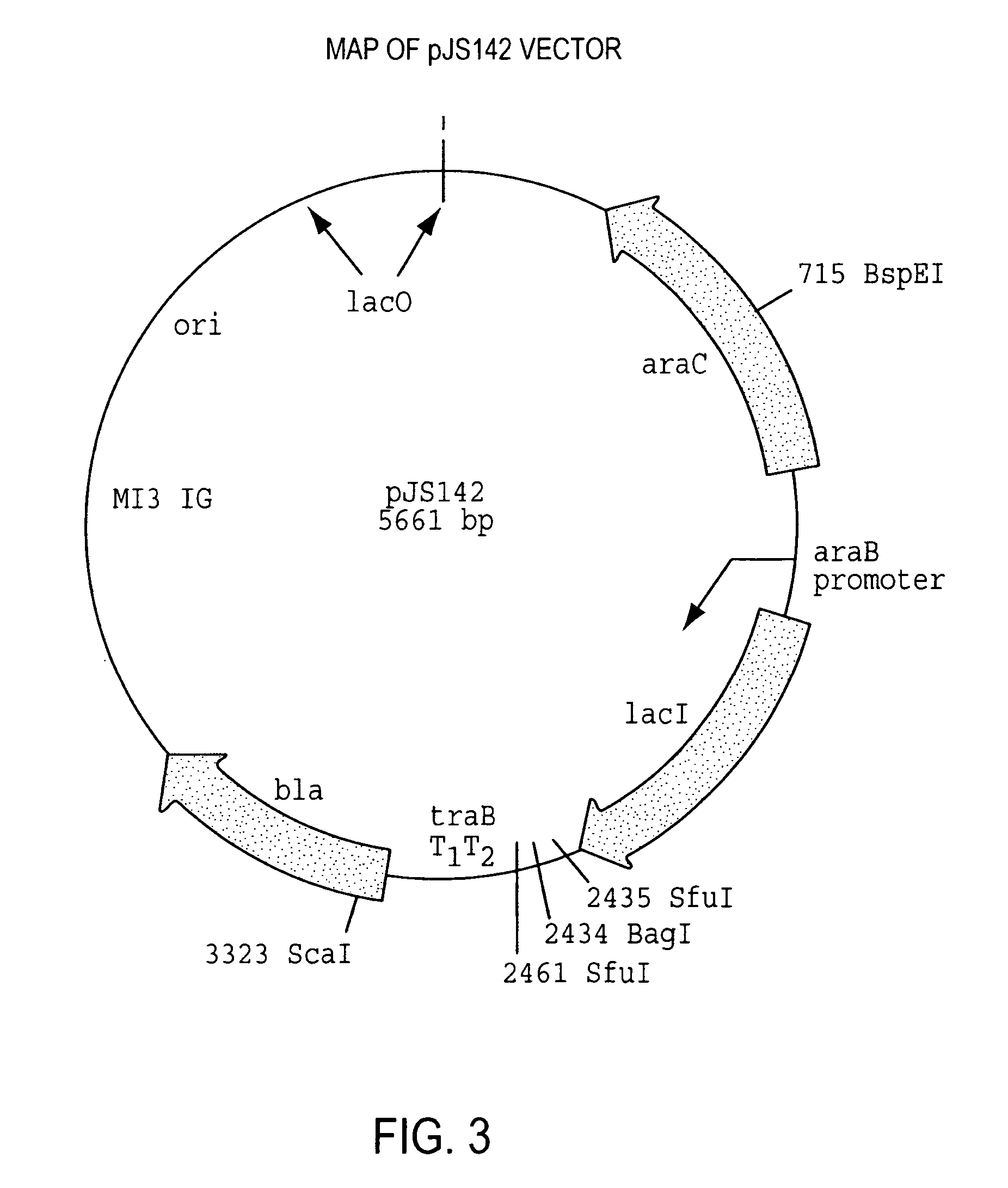
Method for identifying modulators of G protein coupled receptor signaling
a technology modulators, which is applied in the field of modulating the activity of g protein coupled receptors, can solve the problems of inconsistent difficulty in seeing at night, inability to detect objects, and confusion, and achieves the effects of slow recovery and confusion
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Construction of a Peptide Library
[0184] Construction of a biased peptide library has been described previously. Martin et al., J. Biol. Chem. 271:361-366, 1996; Schatz et al., Meth. Enzymol. 267:171-191, 1996. The vector used for library construction was pJS142 (see FIG. 3). This vector had a linker sequence between the LacI and the biased undecamer peptide coding sequence, as well as restriction sites for cloning the library oligonucleotide. The oligonucleotide synthesized to encode the mutagenesis library was synthesized with 70% of the correct base and 10% of each of the other bases at each position. This mutagenesis rate leads to a biased library such that there is approximately a 50% chance that any of the 11 codons will be the appropriate (native) amino acid and approximately a 50% chance that it will be another amino acid. In addition, a linker of four random NNK (where N denotes A, C, G or T and K denotes G or T) codons were synthesized at the 5′ end of the sequence to make...
example 2
Sequences for the Creation of Gα Subunit Peptide Libraries
[0185] Libraries were created using the methods of Example 1 and the sequences listed below in Table VII.
TABLE VIIC-Terminal Gα Subunit Peptide Library Constructs.GαSEQSub-IDunitRELinkerPeptide Coding RegionStopRENO:Gs5-GAGGTGGTNNKNNKNNKNNKattcgtgaaaacttaaaagattgtggtcgtttcTAACTAAGTAAAGC-3′14G115-GAGGTGGTNNKNNKNNKNNKctgcagctgaacctgaaggagtacaatctggtcTAACTAAGTAAAGC-3′119G125-GAGGTGGTNNKNNKNNKNNKctgcaggagaacctgaaggacatcatgctgcagTAACTAAGTAAAGC-3′120G135-GAGGTGGTNNKNNKNNKNNKctgcatgacaacctcaagcagcttatgctacagTAACTAAGTAAAGC-3′121G155-GAGGTGGTNNKNNKNNKNNKctcgcccggtacctggacgagattaatctgctgTAACTAAGTAAAGC-3′122Gz5-GAGGTGGTNNKNNKNNKNNKatacagaacaatctcaagtacattggcctttgcTAACTAAGTAAAGC-3′123
example 3
Isolation of Membranes from Insect Cells Expressing Thrombin Receptor
[0186] Sf9 cells (2×108 cells) were cultured with 200 ml of Grace's insect cell culture medium (Life Technologies, Inc., Grand Island, N.Y.) containing 0.1% Pluronic F-68 (Life Technologies, Inc., Grand Island, N.Y.)), 10% fetal calf serum, and 20 μg / ml gentamicin in a 1-liter spinner flask at 27° C. for 25 hours. Sf9 cells were infected with the ThR / pBluebac recombinant virus at a multiplicity of infection of 3-5, and cultured at 27° C. for 4 days. The cells were harvested, washed with phosphate buffered saline, and then resuspended in 10 mM Tris-HCl, pH 7.4. Cells were then homogenized with a hand-held homogenizer set at low speed for 20 seconds. The broken cells then were sedimented at 17,000×g for 15 minutes. The supernatant was discarded, and the pellet resuspended in a buffer consisting of 50 mM Tris-HCl, pH 7.4 and 10% glycerol. Concentration of receptor in the membrane preparation ranged from 1-10,000 pmol...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular mass | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| excitation wavelength | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com