Nucleic acid amplification in the presence of modified randomers
a technology of randomers and nucleic acids, applied in the field of template dependent polymerase catalyzed primer extension reactions, can solve the problems of inability to control the reproducibility and quality of chemically modified enzyme preparations, time-consuming and inconvenient methods, and unspecific amplification products, etc., to eliminate the inhibition of polymerase, short activation time, and convenient us
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
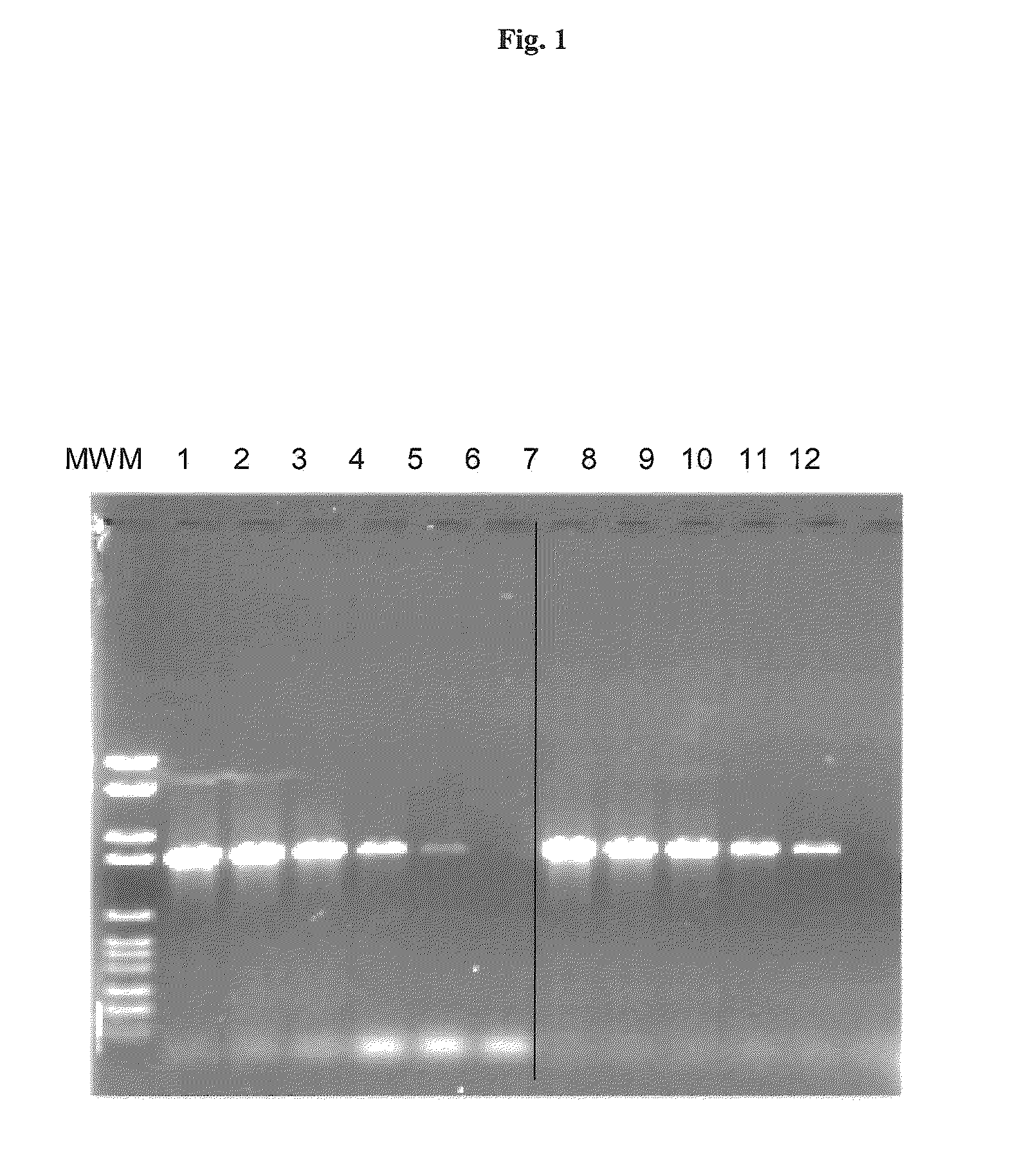
[0128]5′ Pyrene-capped hexamers were analyzed in DNA amplification. PCR reactions in the presence or absence of 100 μM Pyrene-capped hexamers were performed in 50 μl reactions containing 50 ng, 25 ng, 10 ng, 5 ng, 1 ng and 0 ng of human genomic DNA, 30 mM Tris-HCl, pH 8.6, 1.5 mM MgCl2, 50 mM KCl, 0.2 mM dNTP's each, 0.4 μM primers (SEQ ID NO: 1 ATT AGA GAA CCA TGT TAA CAC TAC CG and SEQ ID NO: 2 GAG GTG AAT GAC CAC TGT TTA TTT TC) and 2.5 units Taq DNA polymerase. The following cycle conditions were used: Initial denaturation for 4 min at 94° C. and 35 cycles with 20 seconds denaturation at 94° C., 30 seconds annealing at 62° C., 60 seconds elongation at 72° C. and a final elongation step of 7 min at 72° C. The amplification products were separated on an agarose gel and visualized by ethidium bromide staining.
[0129]The result depicted in FIG. 1 shows a clear improvement in amplification specificity in the presence of pyrene-capped hexamers.
example 2
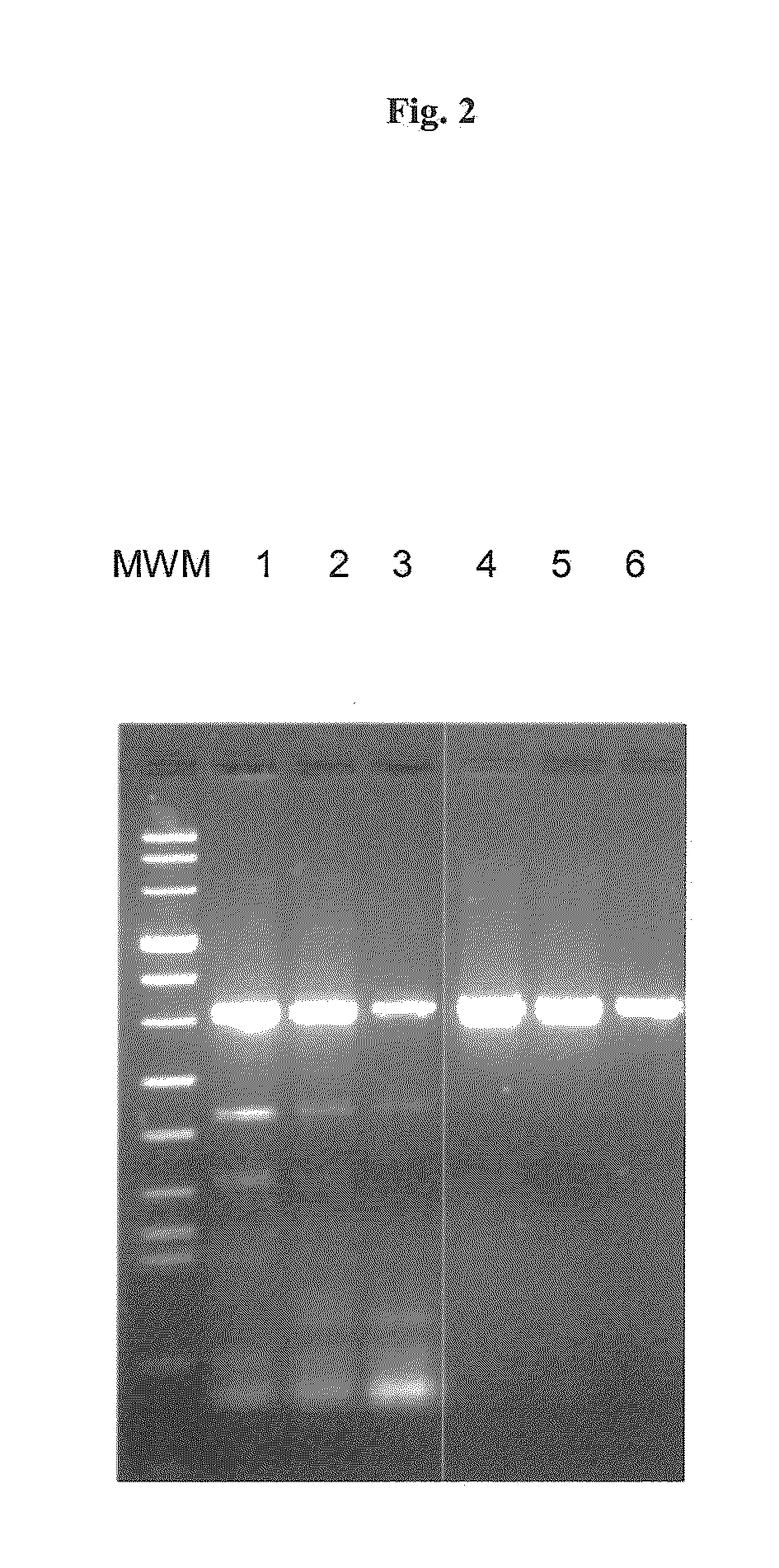
[0130]5′ Pyrene-capped hexamers were analyzed in realtime PCR. PCR reactions in the presence or absence of Pyrene-capped hexamers were performed in 20 μl rections containing 30 ng, 3 ng or 0.3 ng of human genomic DNA, 50 mM Tris-HCl, pH 8.6, 0.2 mM CHAPS, 1 mM BigChap, 20 mM KCl, 3 mM MgCl2, 0.4 μM primers (SEQ ID NO: 3 GGA AGT ACA GCT CAG AGT TCT GC and SEQ ID NO: 4 GAA TCT CCA TTC ATT CTC AAA AGG ACT), 0.2 mM deoxynucleotides, and 2.5 units Taq DNA polymerase. PCR was performed in a LIGHTCYCLER 480 Instrument (Roche Diagnostics GmbH) with the following cycle conditions: Initial denaturation for 2 min at 95° C. and 45 cycles with 1 second denaturation at 95° C., 10 seconds annealing at 65° C. and 10 seconds elongation at 72° C. The amplification products were separated on agarose gel and visualized by ethidium bromide staining (FIG. 2). The result shows a clear improvement in amplification specificity by pyrene-capped hexamers.
example 3
[0131]5′ Pyrene-capped pentamers were analyzed in the same experimental setup as described in example 2. The final concentrations tested were 50 μM, 100 μM, 150 μM and 200 μM. A variety of PCR products were formed in the control reaction (absence of additive), whereas in the presence of increasing amounts of pyrene-capped pentamers the desired product was formed with increased yield. Specificity and sensitivity were significantly higher than in the control experiment without additives (not shown).
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperatures | aaaaa | aaaaa |
| temperatures | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com