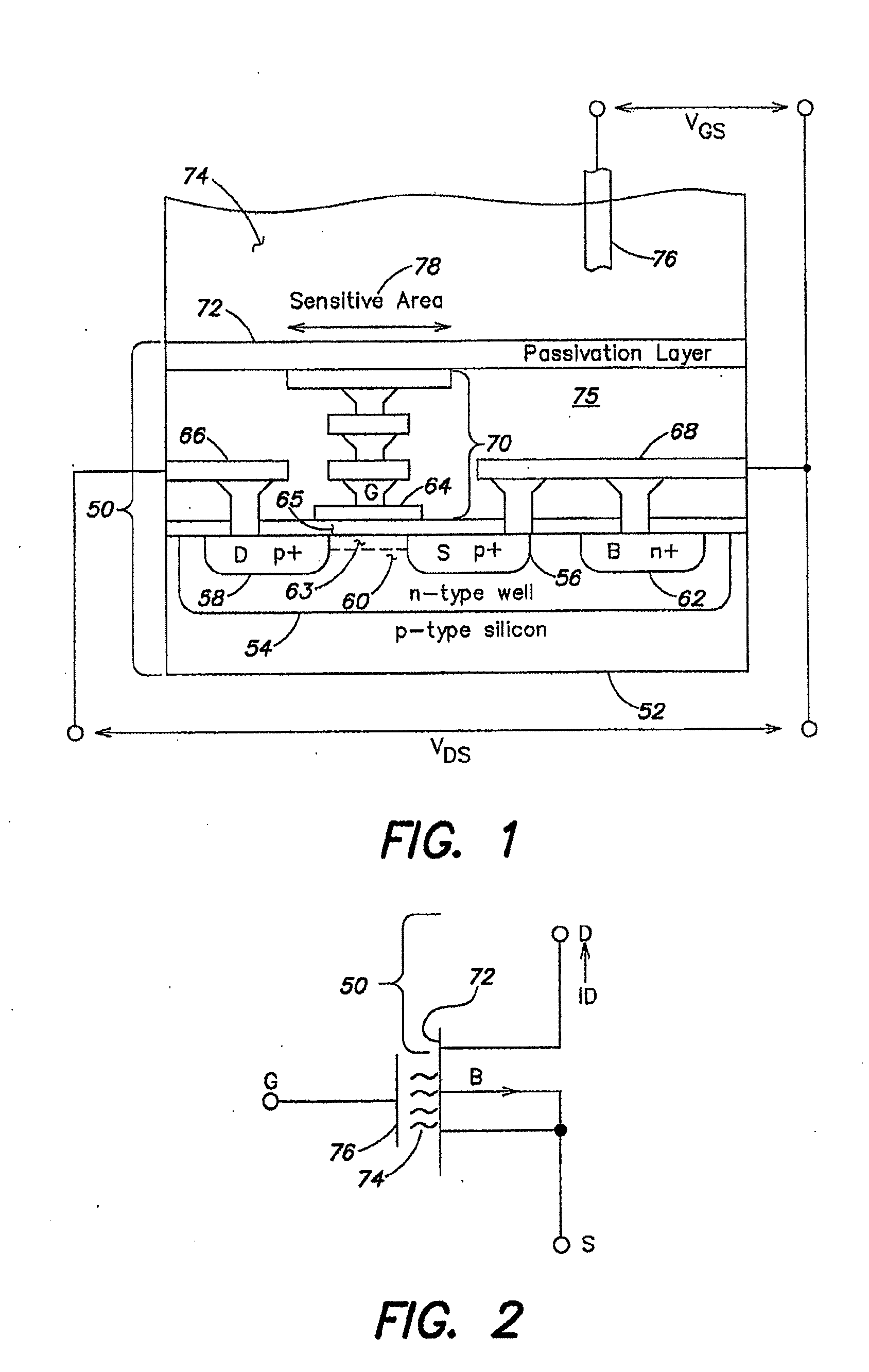
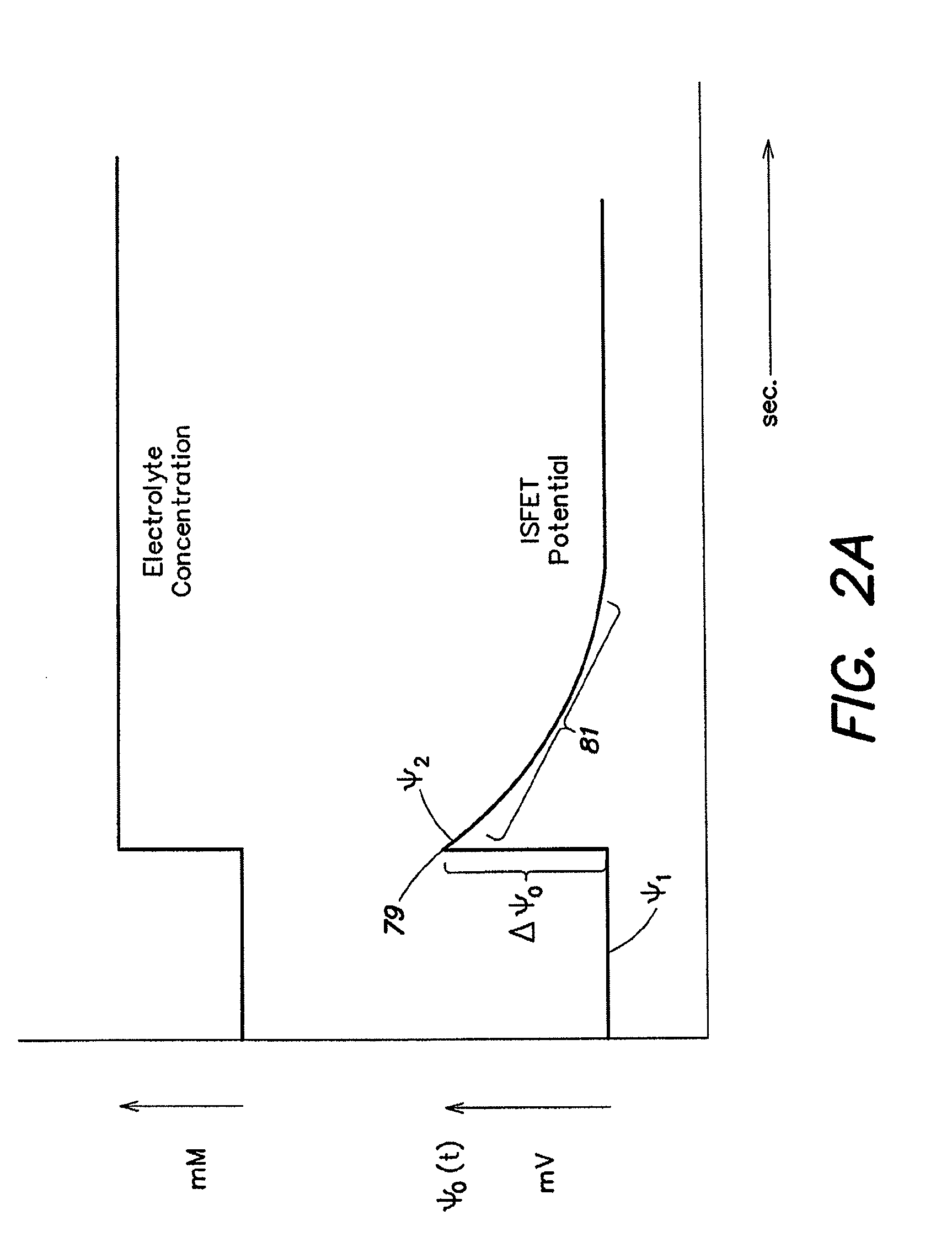
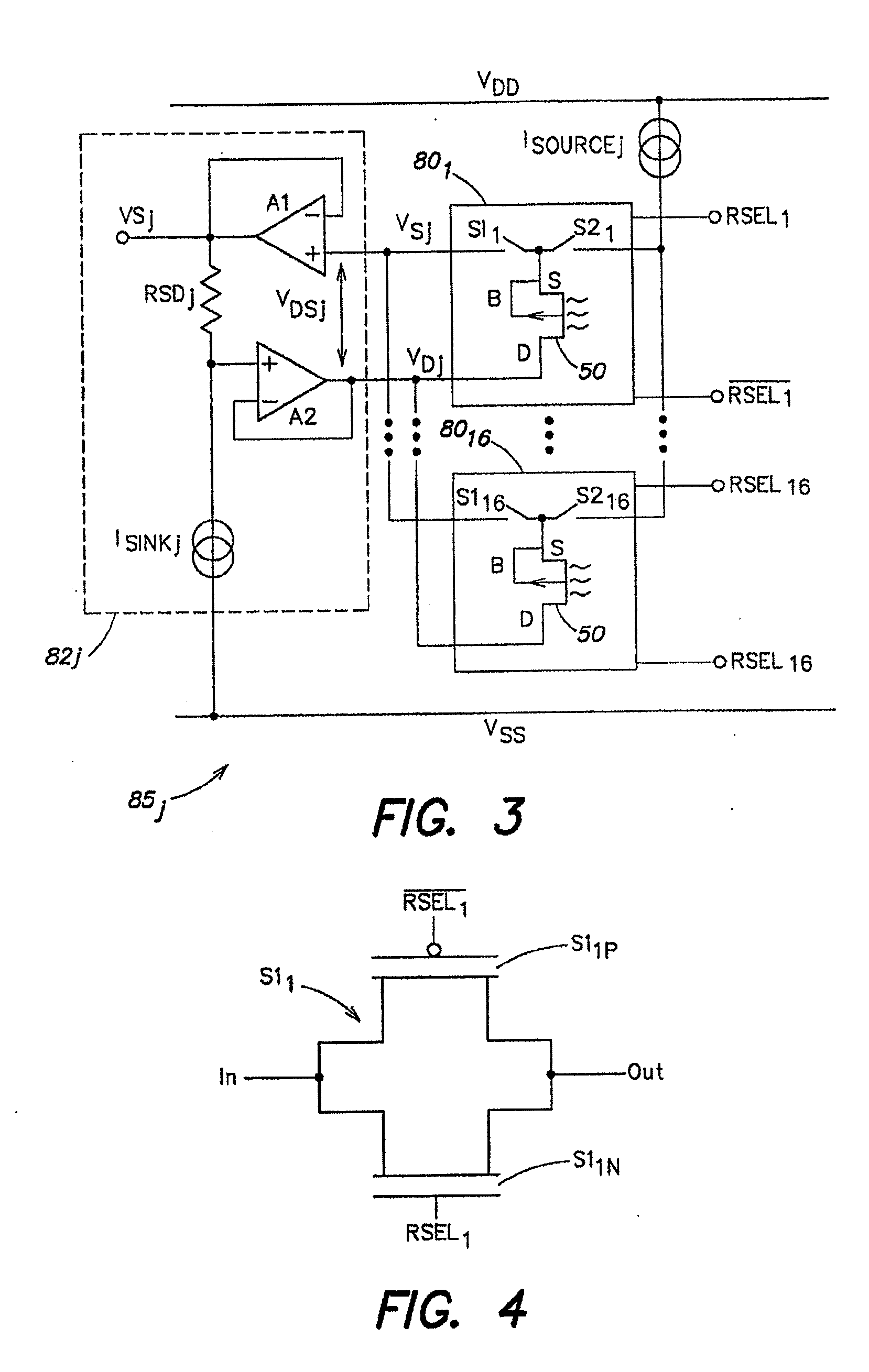
Method and Apparatus for Rapid Nucleic Acid Sequencing
a nucleic acid and sequencing method technology, applied in the field of methods and apparatus for rapid nucleic acid sequencing, can solve the problems of inability to predict the operation of the transmission gate, or to exclude the operation entirely, and the length of individual templates that can be sequenced, and achieve the effects of increasing the number of template nucleic acids used per sensor, and optionally per reaction chamber, and increasing the magnitude of charge chang
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Bead Preparation
[0718]Binding of Single-Stranded Oligonucleotides to Streptavidin-Coated Magnetic Beads. Single-stranded DNA oligonucleotide templates with a 5′ Dual Biotin tag (HPLC purified), and a 20-base universal primer were ordered from IDT (Integrated DNA Technologies, Coralville, Ind.). Templates were 60 bases in length, and were designed to include 20 bases at the 3′ end that were complementary to the 20-base primer (Table 4, italics). The lyophilized and biotinylated templates and primer were re-suspended in TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8) as 40 μm stock solutions and as a 400 μM stock solution, respectively, and stored at −20° C. until use.
[0719]For each template, 60 μl of magnetic 5.91 μm (Bangs Laboratories, Inc. Fishers, Ind.) streptavidin-coated beads, stored as an aqueous, buffered suspension (8.57×104 beads / p L), at 4° C., were prepared by washing with 120 μl bead wash buffer three times and then incubating with templates 1, 2, 3 and 4 (T1, T2, T3, T4: T...
example 2
On-Chip Polymerase Extension Detected by pH Shift on an ISFET Array
[0734]Streptavidin-coated 2.8 micron beads carrying biotinylated synthetic template to which sequencing primers and T4 DNA polymerase are bound were subjected to three sequential flows of each of the four nucleotides. The template sequence downstream of the sequencing primer was a G(C)10(A)10 (SEQ ID NO:5). Each nucleotide cycle consisted of flows of dATP, dCTP, dGTP and dTTP, each interspersed with a wash flow of buffer only. Flows from the first cycle are shown in blue, flows from the second cycle in red, and the third cycle in yellow. As shown in FIG. 72A, signal generated for both of the two dATP flows were very similar. FIG. 72B shows that the first (blue) trace of dCTP is higher than the dCTP flows from subsequent cycles, corresponding to the flow in which the polymerase should incorporate a single nucleotide per template molecule. FIG. 72C shows that the first (blue) trace of dGTP is approximately 6 counts hig...
example 3
Sequencing in a Closed System and Data Manipulation
[0735]Sequence has been obtained from a 23-mer synthetic oligonucleotide and a 25-mer PCR product oligonucleotide. The oligonucleotides were attached to beads which were then loaded into individual wells on a chip having 1.55 million sensors in a 1348×1152 array having a 5.1 micron pitch (38400 sensors per mm2). About 1 million copies of the synthetic oligonucleotide were loaded per bead, and about 300000 to 600000 copies of the PCR product were loaded per bead. A cycle of 4 nucleotides through and over the array was 2 minutes long. Nucleotides were used at a concentration of 50 micromolar each. Polymerase was the only enzyme used in the process. Data were collected at 32 frames per second.
[0736]FIG. 73A depicts the raw data measured directly from an ISFET for the synthetic oligonucleotide (SE ID NO:6). One millivolt is equivalent to 68 counts. The data are sampled at each sensor on the chip (1550200 sensors on a 314 chip) many time...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Volume | aaaaa | aaaaa |
| Distance | aaaaa | aaaaa |
| Mass | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com