Cytosolic isobutanol pathway localization for the production of isobutanol
a technology of isobutanol and cytosol, which is applied in the direction of lyase, enzymology, biofuels, etc., can solve the problems of increasing the demand for domestically produced biofuels, increasing the cost of fuel, and increasing the demand for alternative fuels, so as to increase the cytosolic activity of isobutanol, and reduce or eliminate the ability to targ
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
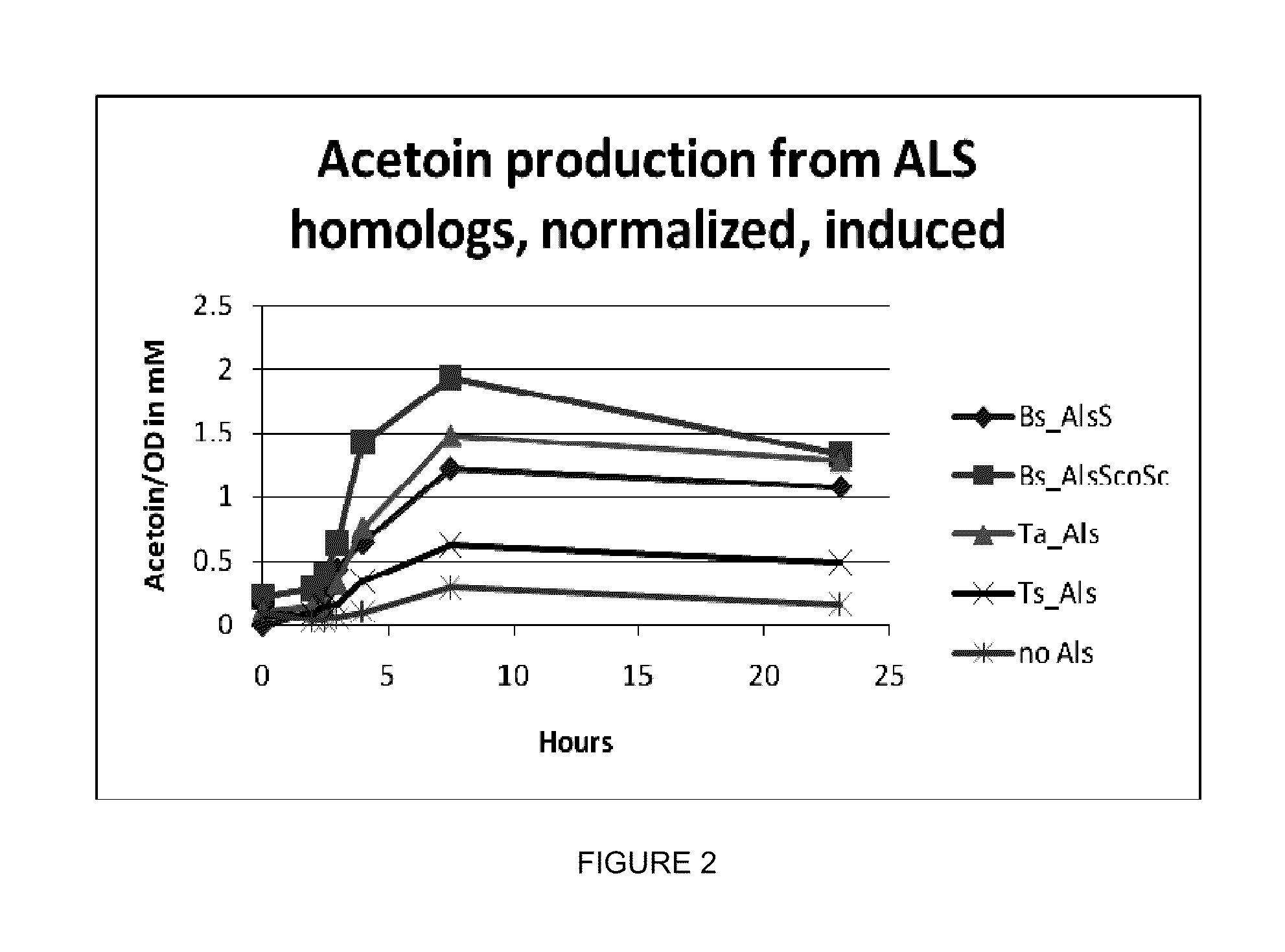
example 1
Isobutanol Pathway is Partially Cytosolic when Expressed in Yeast
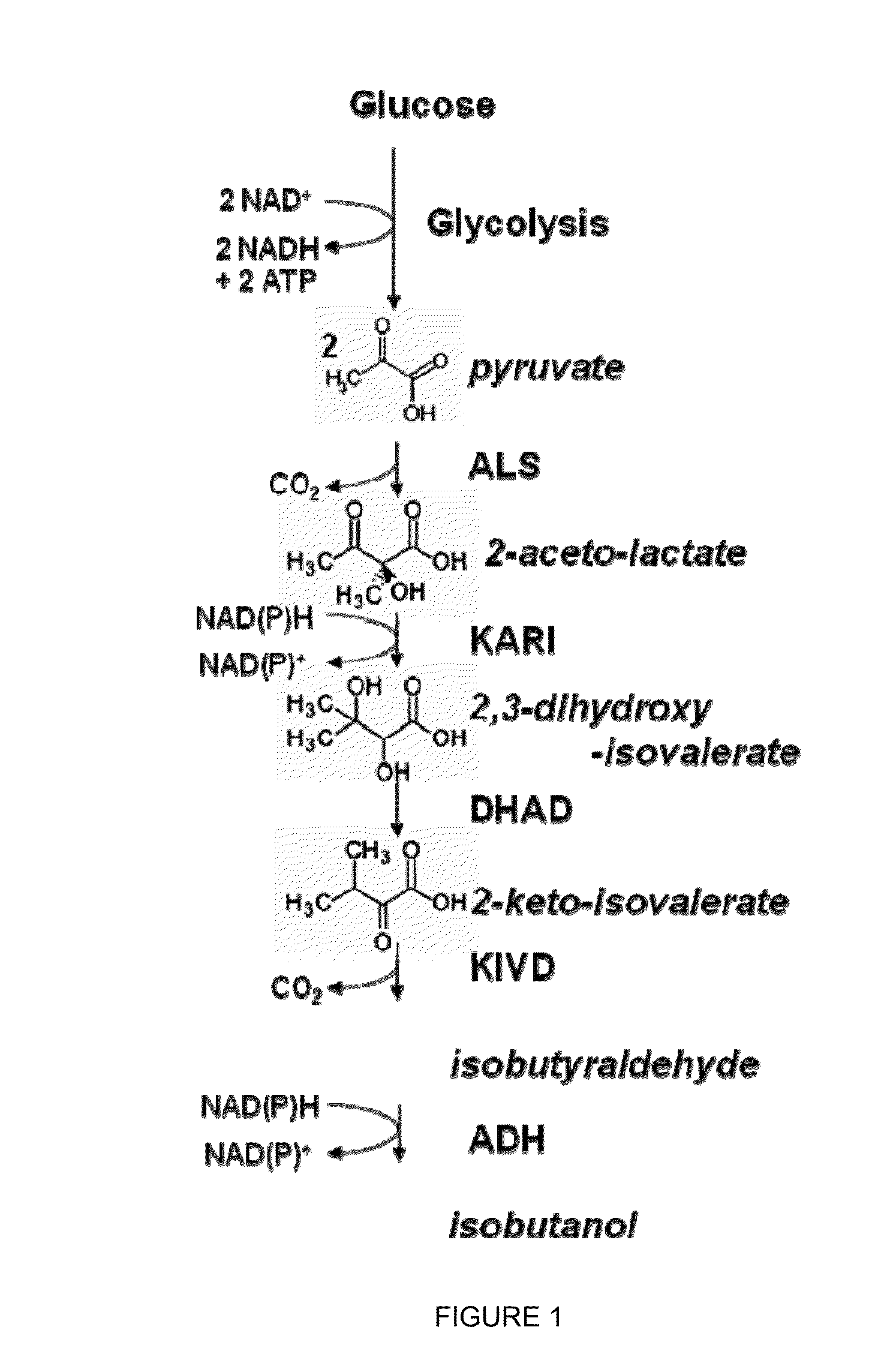
[0273]The purpose of this example is to illustrate that three enzymes in the isobutanol biosynthetic pathway (acetolactate synthase, ketoisovalerate decarboxylase, and isobutanol dehydrogenase) are localized to the cytosol when expressed in yeast.
TABLE 3Genotype of strains disclosed in Example 1.GEVO No.Genotype / Source1287K. lactis ATCC 200826 MAT α uraA1 trp1 leu2 lysA1ade1 lac4-8 [pKD1]1742K. lactis ATCC 200826 MAT α uraA1 trp1 leu2 lysA1ade1 lac4-8 [pKD1] pdc1::KanR1829K. lactis ATCC 200826 MAT α uraA1 trp1 leu2 lysA1ade1 lac4-8 [pKD1] pdc1::kanR {PTDH3:Ec_ ilvC-ΔN;PTEF1:Ec_ilvD-ΔN(codon optimized for K. lactis):ScLEU2 integrated} {PTEF1:Ll_kivD; PTDH3ScADH7:KmURA3 integrated} {PCUP1-1:Bs_alsS, TRP1 randomintegrated}
TABLE 4Plasmids disclosed in Example 1.pGV No.GenotypepGV1503ScTEF1promoter-kanR bla, pUC ori (GEVO)pGV1537KlPDC1 promoter region + Klpdc1 3′UTR sequence,ScTEF1promoter-kanR bla, pUC ori (GEVO)pGV1590TEF...
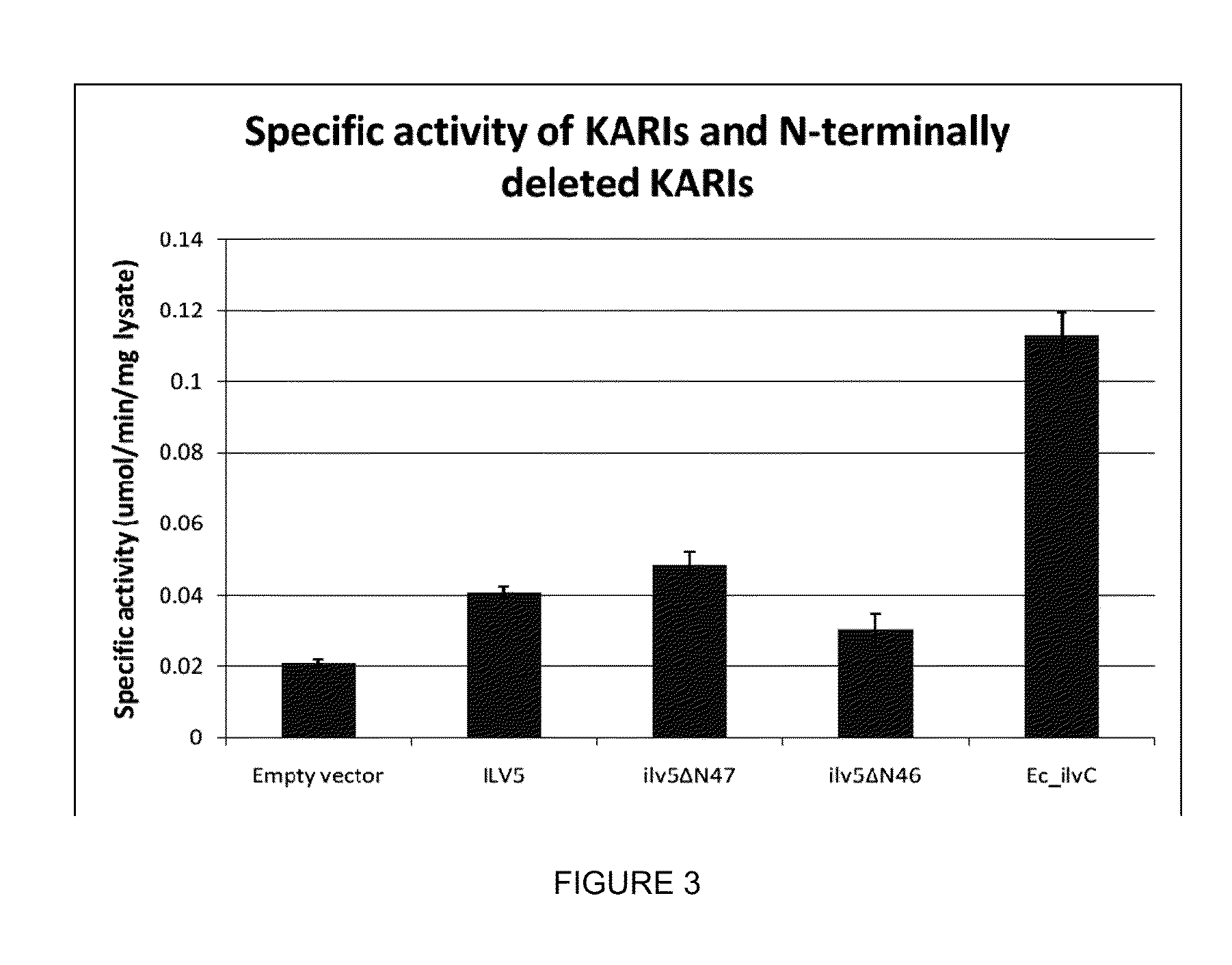
example 2
Construction of an ILV3 Deletion Mutant
[0284]The purpose of this example is to describe the construction of an ILV3 deletion mutant of S. cerevisiae, GEVO2244.
TABLE 8Genotype of strains disclosed in Example 2.GEVO No.Genotype / SourceGEVO1147K. lactis, NRRL Y-1140, (obtained from USDA)GEVO1188S. cerevisiae, CEN.PK, (obtained from Euroscarf);MATα ura3 leu2 his3 trp1GEVO2145S. cerevisiae, CEN.PK; MATα ura3 leu2 his3 trp1ilv3::Kl_URA3GEVO2244S. cerevisiae, CEN.PK; MATα ura3 leu2 his3 trp1 ilv3Δ
TABLE 9Plasmids disclosed in Example 2.Plasmid nameGenotypepUC19bla, pUC-ori (obtained from Invitrogen)pGV1299K. lactis URA3, bla, pUC-ori (GEVO)
[0285]Plasmid pGV1299 was constructed by cloning the K. lactis URA3 gene into pUC19. The K. lactis URA3 was obtained by PCR using primers 575 and 576 from K. lactis genomic DNA. The PCR product was digested with EcoRI and BamHI and cloned into pUC19 which was similarly digested. The K. lactis URA3 insert was sequenced (Laragen Inc) to confirm correct seque...
example 3
DHAD Activity is Localized to Mitochondria
[0290]The purpose of this Example is to demonstrate that the DHAD activity encoded by ScILV3 is localized to the mitochondria.
TABLE 9Genotype of strains disclosed in Example 3.GEVO No.Genotype / SourceGevo2244S. cerevisiae, CEN.PK; MATα ura3 leu2 his3 trp1 ilv3Δ
TABLE 10Plasmids disclosed in Example 3.pGV No.GenotypepGV1106pUC ori, bla (AmpR), 2 micron ori, URA3, TDH3promoter-Myc tag-polylinker-CYC1 terminatorpGV1900pUC ori, bla (AmpR), 2 micron ori, URA3, TEF1promoter-ScILV3(FL)
[0291]Plasmid pGV1106 is a variant of p426GPD (described in Mumberg et al, 1995, Gene 119-122). To obtain pGV1106, annealed oligos 271 and 272 were ligated into p426GPD that had been digested with SpeI and XhoI, and the inserted DNA was confirmed by sequencing.
[0292]Plasmid pGV1900 was generated by amplifying the full-length, native ScILV3 nucleotide sequence from S. cerevisiae strain CEN.PK genomic DNA using primers 1617 and 1618. The resulting 1.76 kb fragment, which ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| ketol-acid | aaaaa | aaaaa |
| dihydroxy acid dehydratase | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com