Ratio based biomarkers and methods of use thereof
a biomarker and ratio technology, applied in the field of ratio-based biomarkers, can solve the problems of erratic tumor response, no simple answer to the question, and the success rate of tumor treatment or tumor treatment after administration of such non-cytotoxic agents is even more complex, so as to reduce the expression of tumor antigen, decrease the expression of pten, and increase the expression of p-akt.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Patient Selection and Tumor Sample Collection
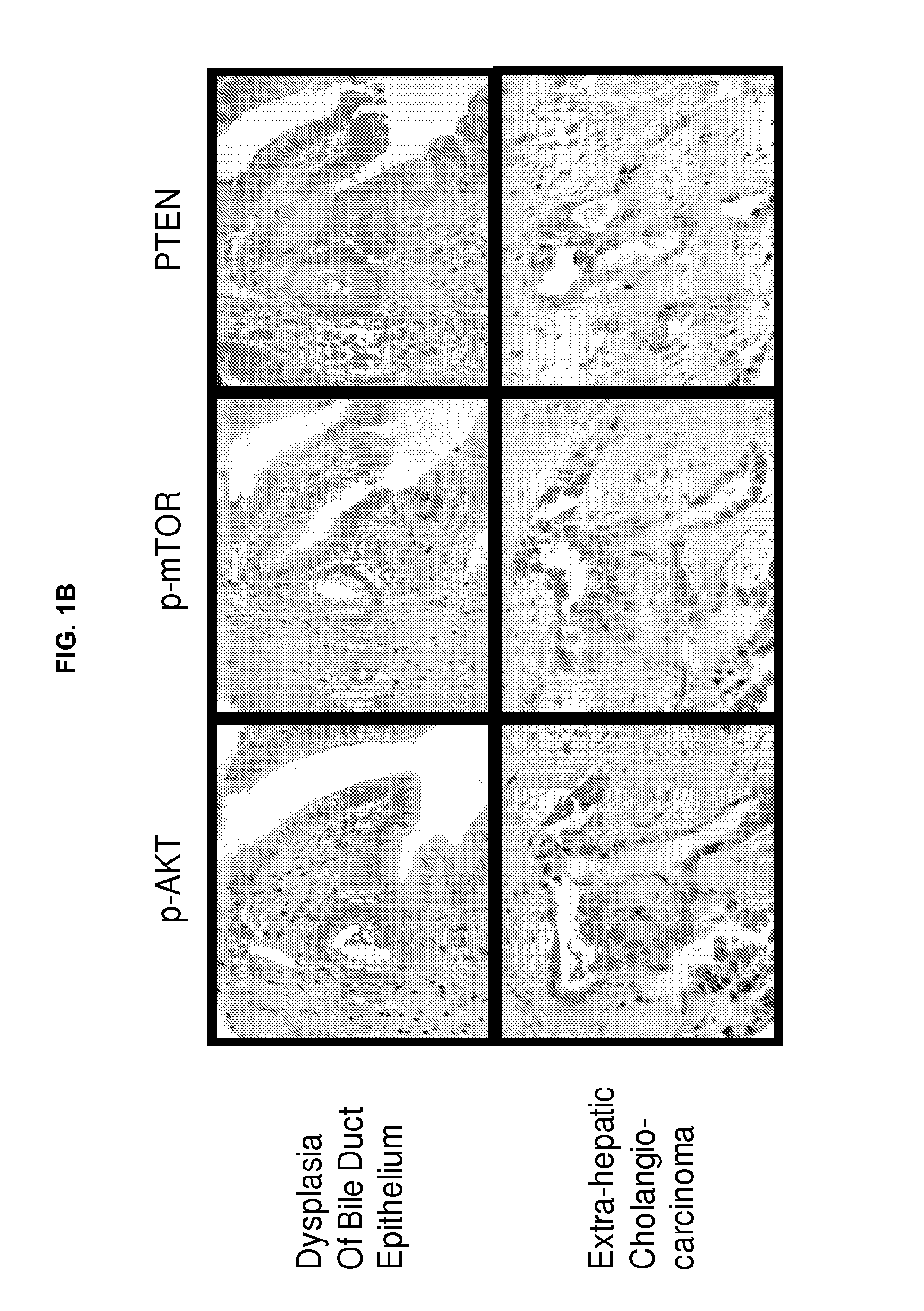
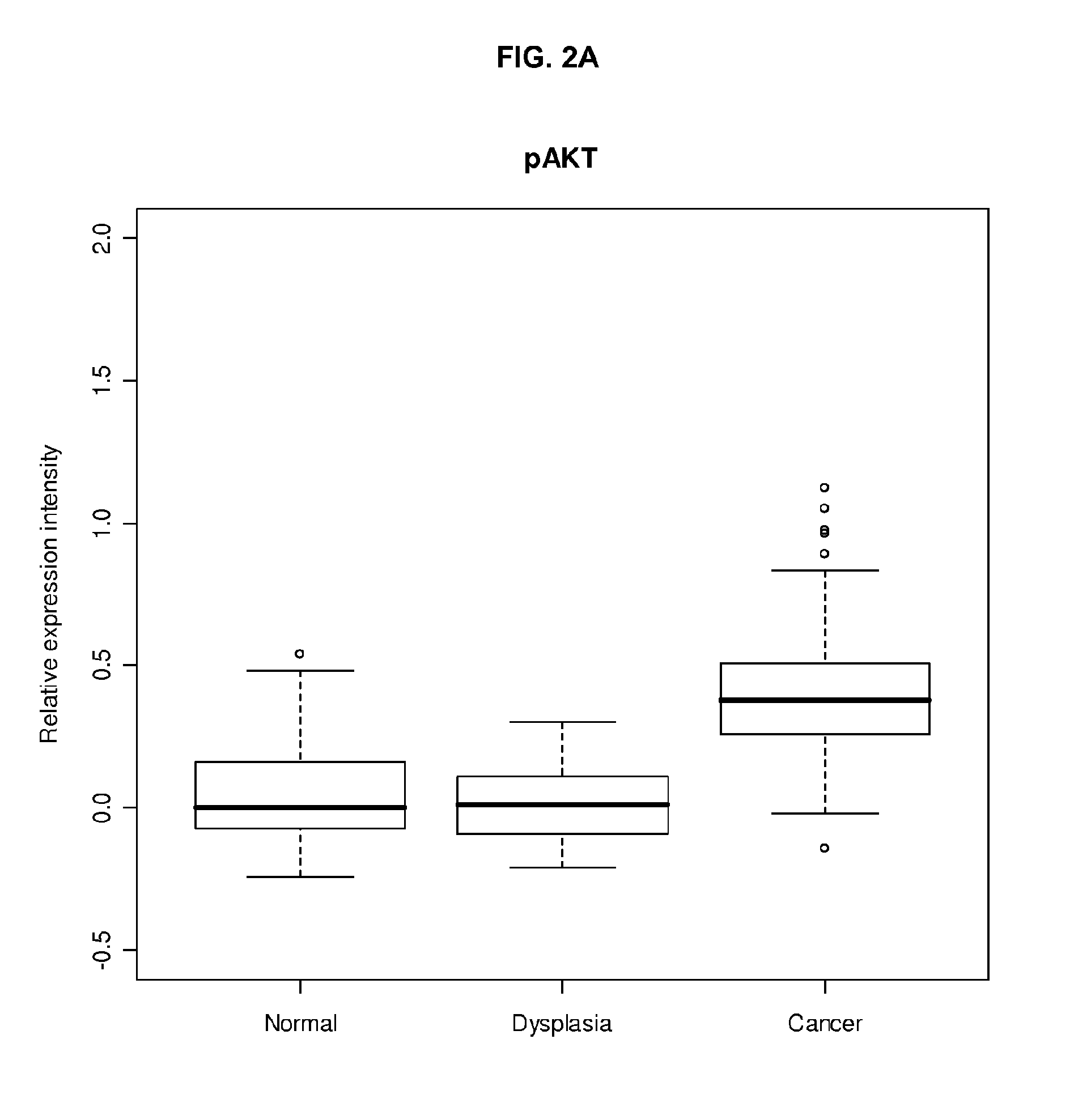
[0308]221 patients with EHCC who were surgically resected at Asian Medical Center, University of Ulsan College of Medicine in Seoul, South Korea were studied. Carcinomas with the epicenter in the extrahepatic bile duct were included, while carcinomas of the ampulla of Vater or pancreas, and those with obvious precancerous epithelial changes in the ampulla of Vater or pancreas were excluded. Carcinomas arising in the gallbladder, or intrahepatic bile duct with extension to the extrahepatic bile duct were also excluded in this study. Medical records were reviewed to obtain data including age and gender of patients, surgical procedure, survival time, and survival status. Data with tumor location, size, and growth pattern were obtained from reviewing pathology reports. Information on post-operative radiation and / or chemotherapy, and performance status of patients was not available for analysis. Material was obtained with appropriate human pro...
example 2
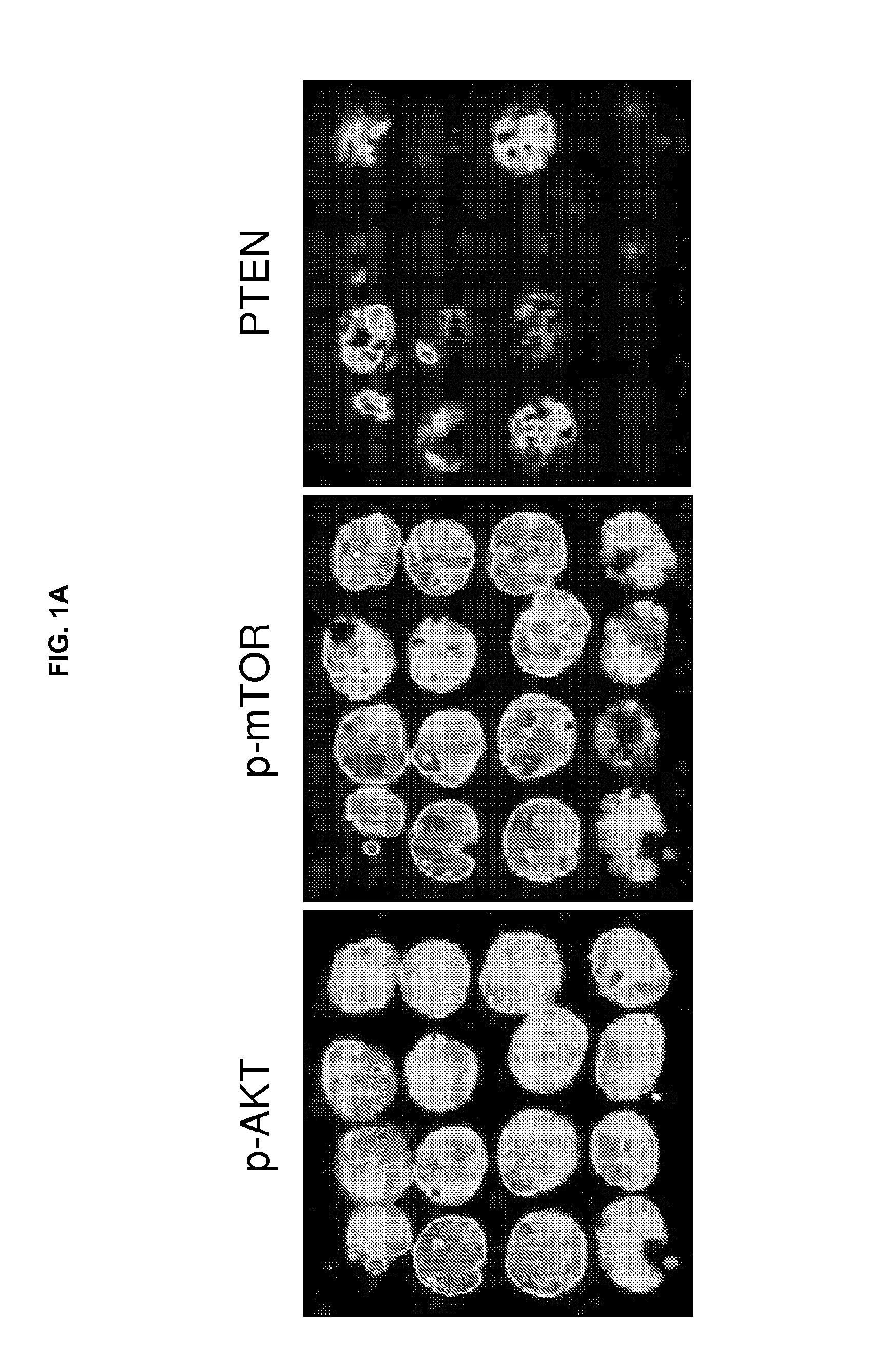
Proteomic Expression Profiling by Multiplex Tissue Immunoblotting
[0311]Multiplex tissue immunoblotting (MTI) was performed as known in the art (for example, Chung et al., Proteomics. 6:676-74, 2006 and Chung et al., Cancer Epidemiol. Biomarkers. Prev. 15:1403-08, 2006). In brief, TMA slides were deparaffinized and treated with an enzyme cocktail solution [0.001% trypsin plus 0.002% proteinase-K, 10% glycerol, 50 mM NH4HCO3 pH 8.2 (Fisher Scientific, Hampton, N.H.)] for 30 minutes at 37° C. Slides were subsequently incubated with Probuffer complete protease inhibitor solution [0.5 ml phosphatase inhibitor I (Sigma, St. Louis, Mo.), 0.5 ml phosphatase inhibitor II (Sigma), 1 protease inhibitor tablet (Roche Diagnostics, Indianapolis, Ind.) in 50 ml PBS (pH 7.2)] for 20 minutes at room temperature (RT). The proteins of treated slides were transferred to a 5-membrane stack (set) of P-FILM (20 / 20 GeneSystems, Rockville, Md.) using Tris-glycine transfer buffer (50 mM Tris, 380 mM Glycine)...
example 3
[0314]Tissue sections were deparaffinized and hydrated in xylene and serial alcohol solutions, respectively. Endogenous peroxidase was blocked by incubation in 3% H2O2 for 10 minutes. Antigen retrieval was performed in a steam pressure cooker with prewarmed antigen retrieval buffer pH 10 (Dako, Glostrup, Denmark) at 95° C., for 10 minutes. To minimize non-specific staining, sections were incubated with protein block (Dako) for 15 minutes. Primary antibodies were incubated overnight at 4° C. Antigen-antibody reactions were detected with DAKO LSAB+ peroxidase kit and DAB. Anti-p-AKT, anti-p-mTOR, and PTEN antibodies (Cell Signaling) were used at a dilution of 1:200. Immunostained sections were lightly counterstained with hematoxylin, dehydrated in ethanol, and cleared in xylene.
PUM
| Property | Measurement | Unit |
|---|---|---|
| depth | aaaaa | aaaaa |
| depth | aaaaa | aaaaa |
| freezing temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com