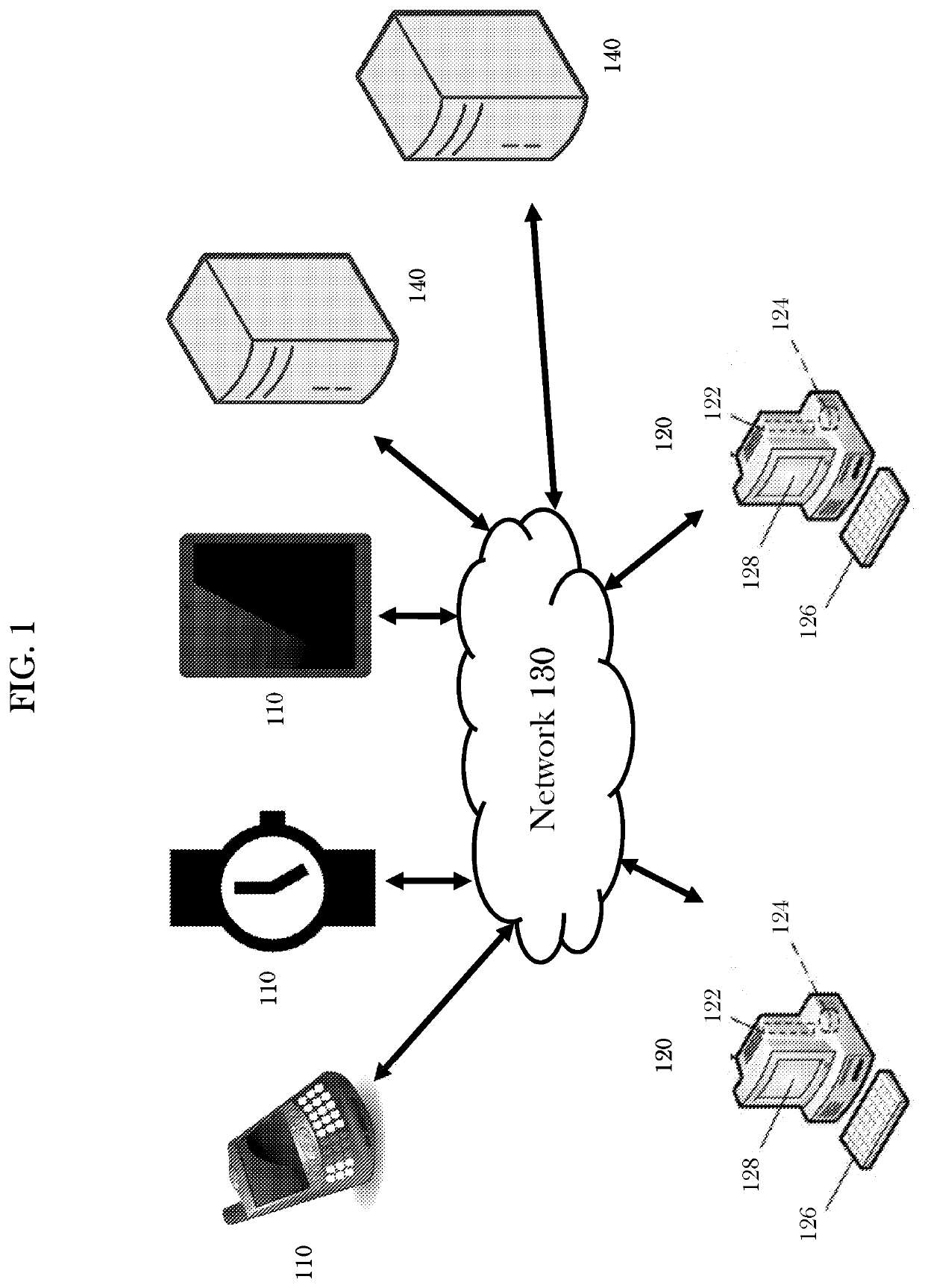
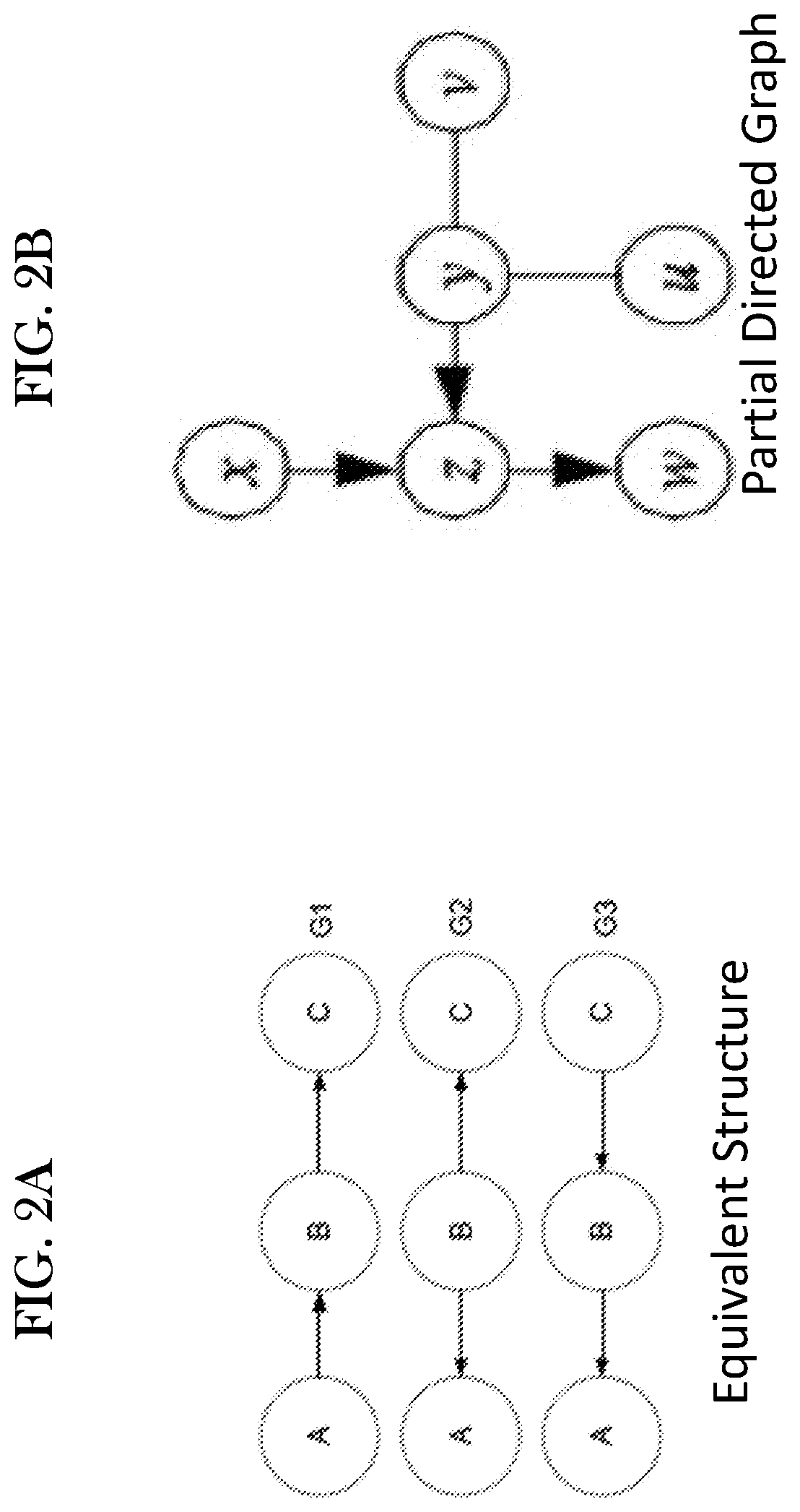
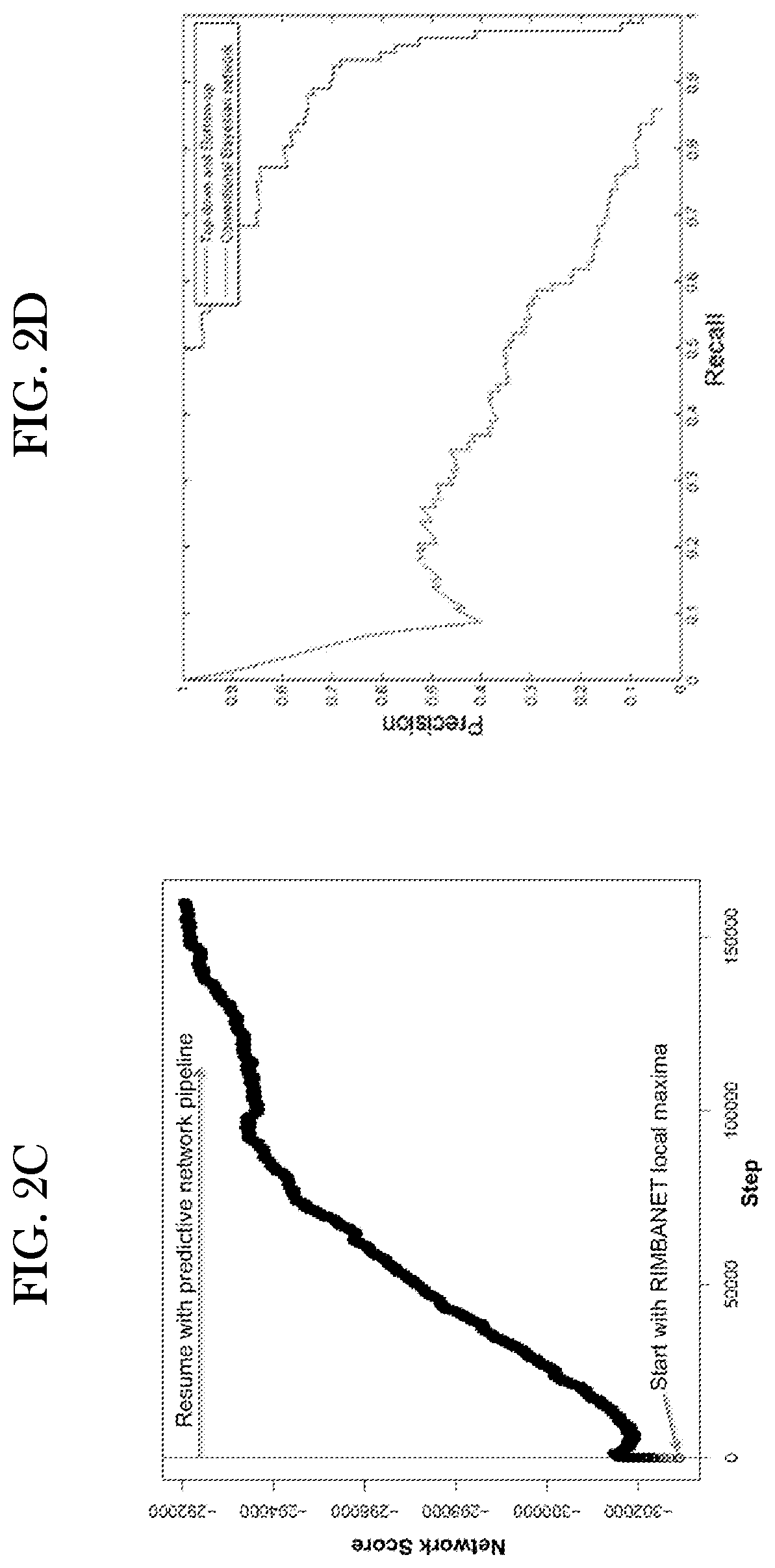
Systems and methods for predictive network modeling for computational systems, biology and drug target discovery
a network modeling and computational system technology, applied in the field of predictive network modeling, can solve the problems of increasing cardiovascular disease risk and their contribution to explain the mechanisms leading to the decrease of insulin sensitivity, and achieve the effects of enhancing bayesian network learning and accurate prediction of the marginal probability of variables
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Predictive Network Analysis Identifies HSPA2 as a Novel Alzheimer's Disease Target
[0093]It is believed that changes in gene and protein expression are crucial to the development of late onset Alzheimer's disease (LOAD). Herein, proteins are examined and incorporated into networks in two separate series, and the outputs are evaluated in two different cell lines. The pipeline included the following steps: (1) Predicting expression quantitative trait loci (eQTLs); (2) Determining differential expression; (3) Analyzing networks of transcript and peptide relationships; and (4) Validating effects in two separate cell lines. We performed all the analysis in two separate brain series to validate effects. The two series included 345 samples in the first set (177 controls, 168 cases; age range 65-105; 58% female; KRONOSII cohort) and 409 samples in the replicate set (153 controls, 141 cases, 115 MCI; age range 66-107; 63% female; RUSH cohort). The top target is Heat Shock Protein Family A Mem...
example 2
Insulin Sensitivity Measurement And IPSC Generation
[0108]The systems and methods of the present invention are also applicable to predictive network modeling in human induced pluripotent stem cells to identify key driver genes for insulin responsiveness.
[0109]Individuals in the study have accompanying genome-wide genotyping and gold standard measurement of insulin sensitivity (i.e. steady state plasma glucose-SSPG-derived from an insulin suppression test). Other biometric parameters include age, body mass index, sex and race / ethnicity (Table 1). Three to seven iPSC lines were generated from each individual, with no apparent differences in the reprogramming efficiency between IR and IS cells. The complete pipeline for iPSC generation and quality control has been previously described. Briefly, RNA-seq data for 317 ipSC lines from 101 individuals was generated and, after quality control, RNA-seq data from 310 samples from 100 individuals was analyzed, of which 48 were IS (149 samples) a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com