Recombined protein carrier PEA II-HphA of actively carrying exogenic gene, its preparing method and use
A technology for exogenous genes and recombinant proteins, which can be used in recombinant DNA technology, botanical equipment and methods, biochemical equipment and methods, etc., and can solve the problems of short expression time, short expression time, and unclearness of non-viral transfer systems.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
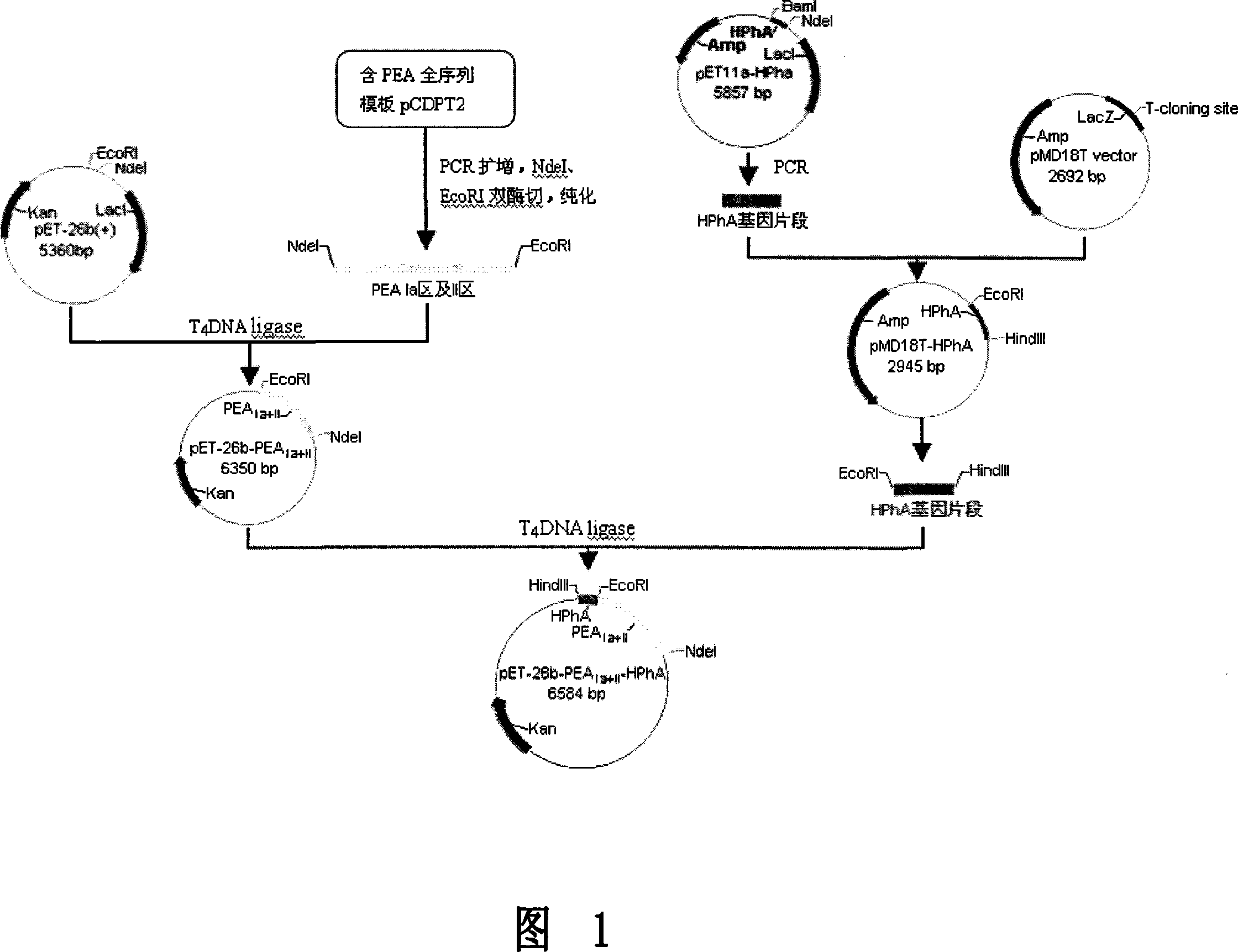
[0060] Example 1: Gene amplification in PEAIa region and region II and ligation with pET26b vector
[0061] The sequences of PEAIa and II regions were searched through GenBank, and amplification primers were designed for them. The upstream primer was marked as P1, and the downstream primer was marked as P2. The NdeI restriction site and the EcoRI restriction site were introduced into the N-terminus of the Ia region and the II region fragment and the C-terminus. pCDPT 2 (Containing PEA whole gene sequence) as template, P1 and P2 are primers to carry out amplification reaction, and the obtained PCR product is marked as PEAIa+II;
[0062] P1 (28bp): 5'GGAATTCCATATGGCCGAGGAAGCCTTC 3' with NdeI restriction site
[0063] P2 (23bp): 5'CGGAATTCCGCGCCGGCCTCGTC 3' with EcoRI restriction site
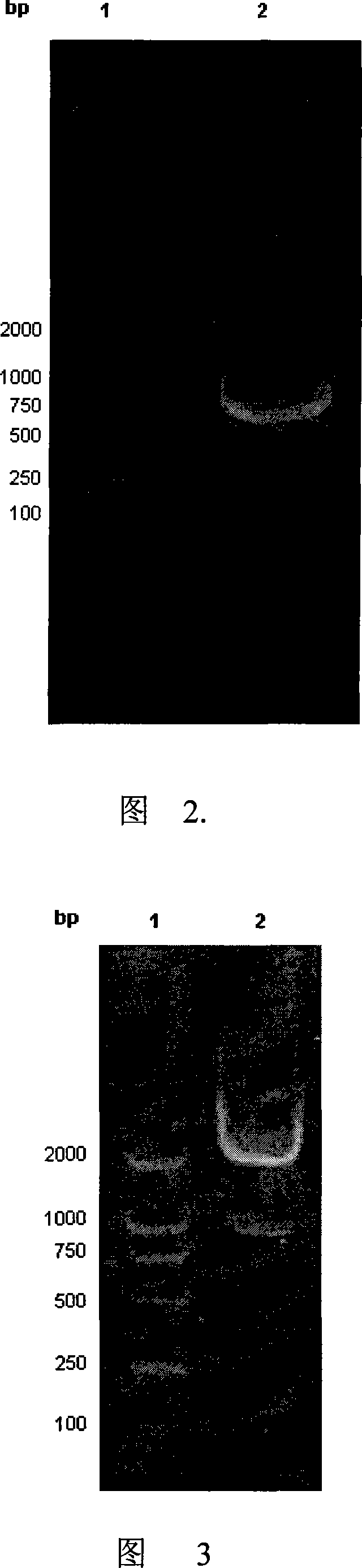
[0064] Due to the high GC content of the gene sequences in PEAIa and II regions, LaTaq enzyme was selected for PCR reaction, and the gene fragments obtained by PCR amplification were analyzed b...
Embodiment 2
[0066] Example 2: Ligation of recombinant plasmid pET26b-PEA Ia+II to HPhA
[0067] The entire gene sequence of Pyrococcus horikoshii OT3 has been determined, which contains the histone gene PHS051. According to the sequence of the gene, two primers with restriction sites are designed to amplify the target gene; the upstream primer is labeled P3, and the downstream primer is labeled is P4; and an EcoR I restriction site and a HindIII restriction site are introduced at the N-terminus of HPhA and the C-terminus of HPhA; the plasmid pET11a-HPhA is used as the template, and P3 and P4 are used as primers to carry out the amplification reaction, and the obtained PCR product is marked as HPhA ;
[0068] P3 (26bp): 5'CGGAATTCGTGTGGATGATGGGAGAA 3' contains EcoR I restriction site
[0069] P4 (28bp): 5'CCCAAGCTTTCAAGCTCTTAATAGCGAGC 3' with HindIII restriction site
[0070] Amplify the gene fragment of HPhA by conventional PCR method, and analyze it by agarose gel electrophoresis.
[...
Embodiment 3
[0072] Example 3: SDS-PAGE results and immunoblotting analysis of recombinant protein PEAIa+II-HPhA expression products
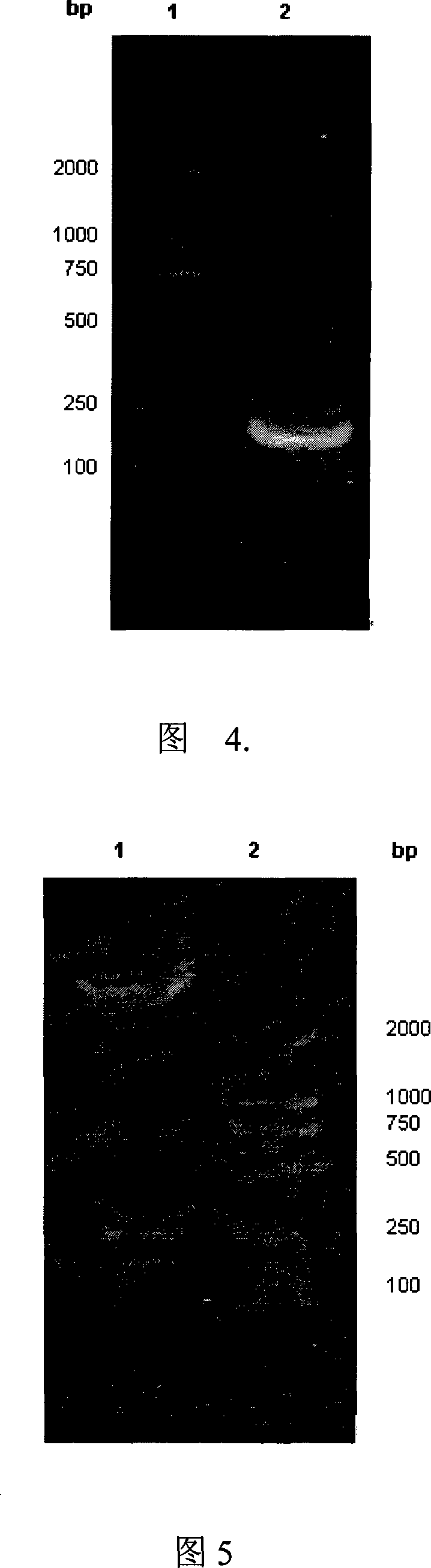
[0073] The recombinant plasmid pET26b-PEAIa+II-HPhA was transformed into the expression bacteria-Escherichia coli BL21-Codon Plus, after induction by IPTG, the SDS-PAGE results showed that the recombinant protein PEA-HPhA has an obvious expression band at about 48.98KD. Values are consistent; see Figure 6;
[0074] The lysate supernatant and precipitate of recombinant protein expressing bacteria were subjected to SDS-PAGE electrophoresis at the same time. The results showed that most of the target protein was in the supernatant, and a small amount of target protein also existed in the corresponding position in the precipitate. It can be expressed in a soluble form under an inducing environment.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com