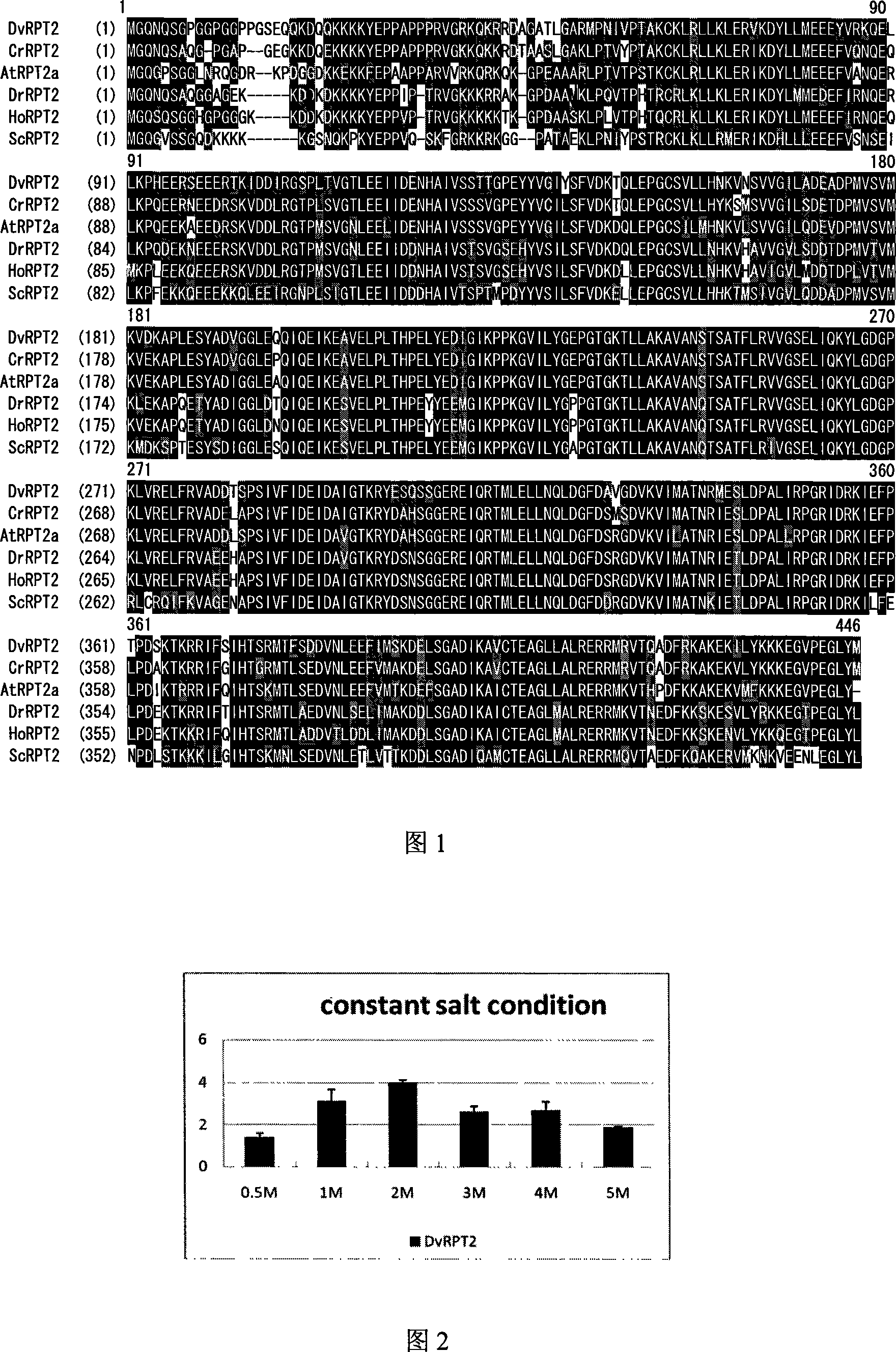
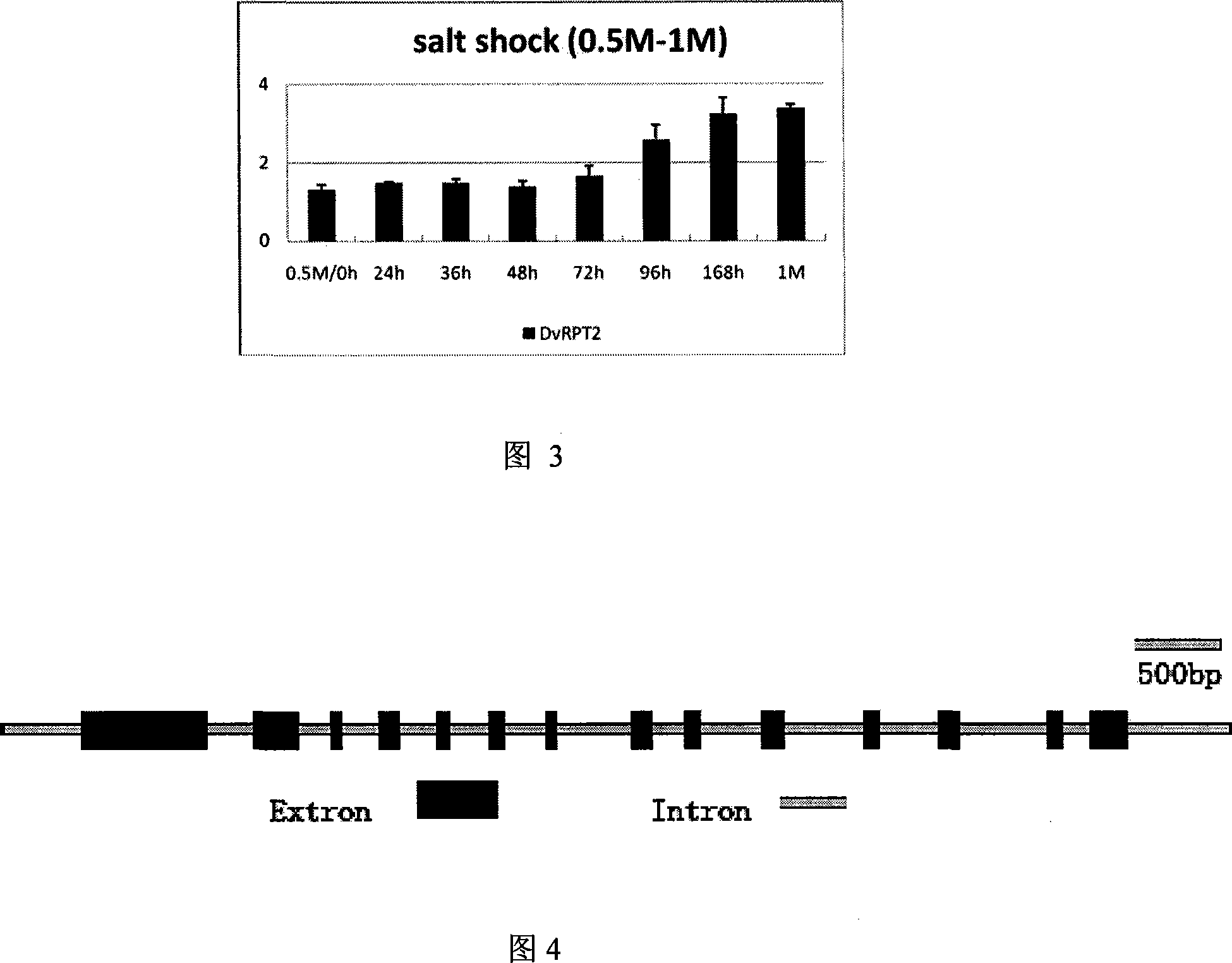
cDNA order of salt algae 26s proteasome subunit RPT2 gene, its coding protein and full-length gene order and clone method
A proteasome, full-length gene technology, applied in the field of cDNA sequence, can solve the problem that the precise functions of the six RPT subunits are not very clear.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Example 1: Cloning and Analysis of DvRPT2 Full-length Coding Region Sequence
[0028] The gene DvRPT2 cloned in this experiment was derived from the Salina EST library in our laboratory. The DvRPT2-containing Salina EST library plasmid was digested with restriction endonucleases (EcoR I / Xho I) to release the target fragment and be used in subsequent experiments. According to sequencing analysis, the longest cDNA fragment of DvRPT2 is 1534bp, which contains the specific tailing signal TGTAA and poly(A) structure of the green algae gene. The analysis results of Vector NTI software showed that the longest reading frame of DvRPT2 encoded a protein containing 446 amino acids, the molecular weight of the protein was 49.7kDa, and the isoelectric point was 5.98. It can be seen that the protein encoded by DvRPT2 is an acidic protein.
Embodiment 2
[0029] Example 2: Sequencing and splicing of DvRPT2 gene sequence
[0030] The BAC clone containing the DvRPT2 gene was obtained by screening the Salina BAC library. Using the Large-Construct Kit of QIAGEN Company to extract the BAC plasmid without Escherichia coli genome contamination, the recovered DNA was mechanically crushed, and the DNA fragments with a size of 2-4 kb were recovered and connected to the pUC18 vector to construct a shotgun library. The primer sequence of the screening library is:
[0031] RPT2P2a: 5′ CCCTGTGAGACTGACTTGAGA 3′
[0032] RPT2P2b: 5' AGCTTGCACTTGGCTGTG 3'
[0033] Large-throughput extraction of plasmid DNA from the shotgun library was performed on the MegaBACE 4500 for sequence determination. Each shotgun plasmid is sequenced bidirectionally using M13 forward and reverse primers. A total of 384 shotgun library plasmids, namely four 96-well plates, were sequenced.
[0034]In the parallel computer group of the RedHat Linux platform, the meas...
Embodiment 3
[0035] Example 3: Expression of DvRPT2 induced by different salt concentrations and 3M salt
[0036] For the study of DvRPT2 at different steady-state salt concentrations, we cultured Salina cells in Salina media containing 0.5, 1M, 2M, 3M, 4M and 5M NaCl at 2×10 6 Cells were collected at the time of cell / ml, and the total RNA of Salina cells was extracted respectively. The above RNA samples were digested with DNase I, reversed into cDNA under the action of reverse transcriptase, diluted 10 times and used as templates for realtime PCR under various experimental conditions. PCR primers:
[0037] RPT2P2a: 5'-GGAGGAACGCACCAAGAT-3'
[0038] RPT2P2b: 5′-CAGGATGCCAACAACAGAGT-3′
[0039] DvActinRT+: 5′-GCCACTGCTTTAGCTGTTTGC-3′
[0040] DvActinRT-: 5′-CCTCATGCTCCCAGATGTTCTA-3′
[0041] After the library plasmids of DvRPT2 and DvACTIN were purified and quantified, the 10-fold serial dilution was used as the standard curve of quantitative PCR, and the transcription amount of DvACTI...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com