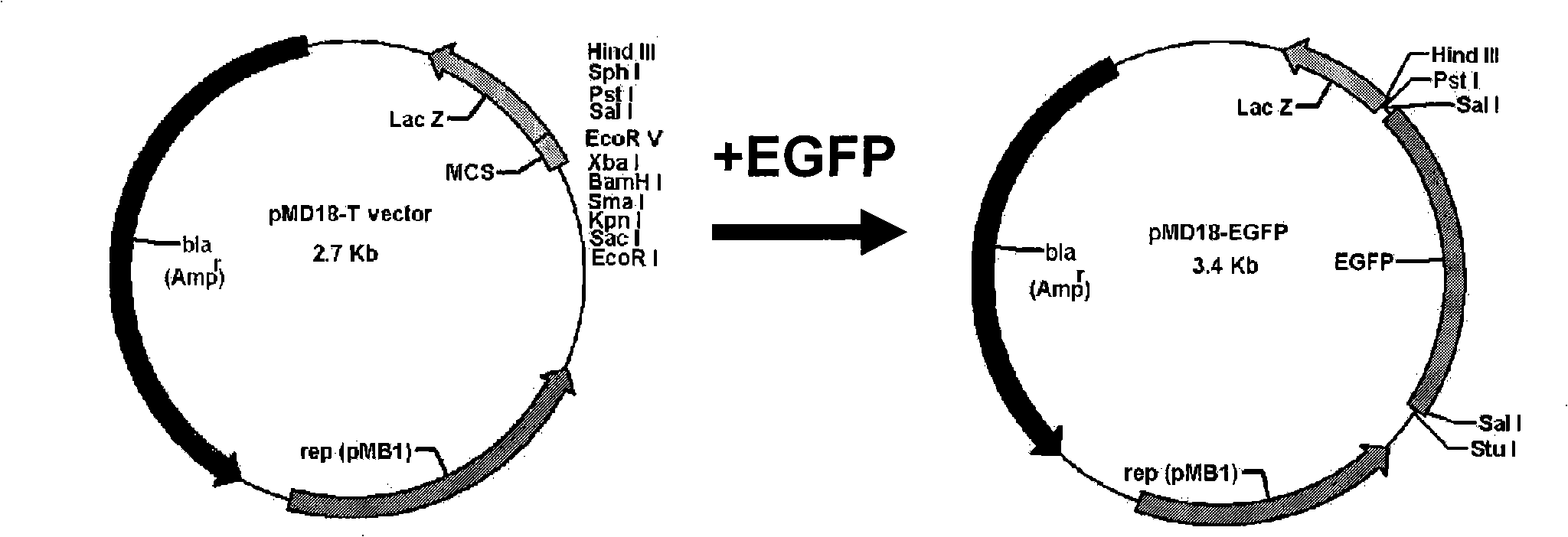
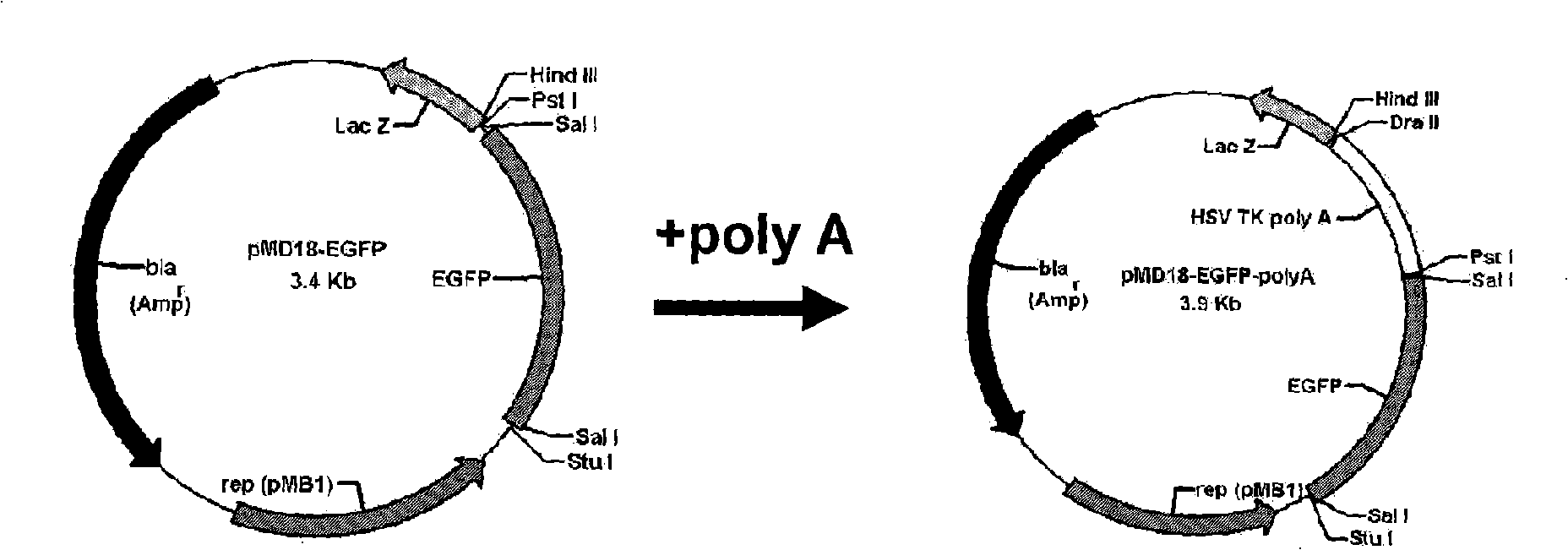
Construction method and application of two-color fluorescence report carrier
A two-color fluorescence and carrier technology, applied in the fields of molecular biology and antiviral research, can solve the problems of inaccuracy, affecting the accuracy of quantification, and the inability to accurately project the inhibition efficiency of siRNA
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
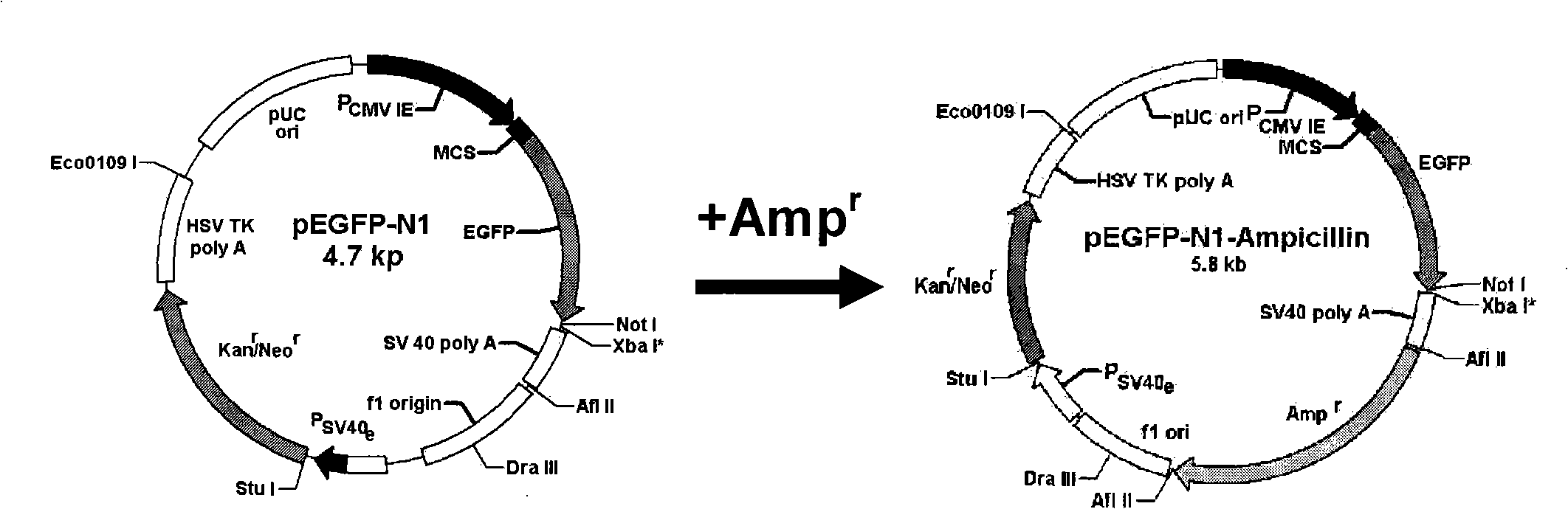
[0096] Embodiment 1PCR amplification has the ampicillin resistance gene sequence of promoter and transcription termination signal
[0097] Synthesis of amplification primers: primers were designed according to the ORF of the ampicillin resistance gene and the vector, and detected by primerpremier 5.0 software (product of Premier Company in Canada). The 5'primer P1 used is: CCTAGAT CTTAAG AAATTAAAAATGAAGTTT (the underline is the introduced Bgl II restriction site); 3'primer P2 is: TAATAATGGTTT CACGTAGTG CAGGTGGC (underlined is the introduced Dra III restriction site). The primers used in the present invention were all synthesized by Wuhan Saibaisheng Company. Using pUC18 (product of Takara Company) as a template, using upstream primers and downstream primers, a DNA sequence from 1566 to 2617 in pUC18 was amplified using a PCR machine.
[0098] PCR system components: 5μl 10×1 # buffer, 5 μl dNTP (2mM), 1 μl primer P1 (10 μM), 1 μl primer P2 (10 μM), 0.1 μl template pUC18, ...
Embodiment 2
[0101] Example 2 Recovery and purification of ampicillin resistance gene PCR product with promoter and transcription termination signal
[0102] (1) Carry out electrophoresis (1×TAE) with 1.0% agarose gel to the PCR product among the embodiment 1, observe electrophoresis situation with ultraviolet light, when the DNA band to be recovered is completely separated from other bands, stop electrophoresis, in Cut off the band to be recovered with a razor blade under ultraviolet light, and purify it with a PCR product purification kit. (2) Crush the glue in an Eppendorf tube, add an equal volume of sol solution Binding Buffer, bathe in water at 65°C for 7 minutes, and shake the Eppendorf tube every 2 minutes until the glue is completely melted. (3) Add the melted sample to the chromatographic column, centrifuge at 12000 rpm for 1 min, and discard the liquid. (4) Add 300 μl BindingBuffer, centrifuge and discard the liquid. (5) Add 750μl Washing Buffer, centrifuge and discard the liq...
Embodiment 3
[0103] Example 3 Plasmid pEGFP-N1 is single-digested by Afl II
[0104] Reaction system: 5 μl 10×0 buffer; pEGFP-N1 plasma 15 μl; 5 μl Afl II enzyme; add sterile water ddH 2 0 to 50 μl.
[0105] Reaction process: Mix and mix the components of the reaction system on ice, centrifuge briefly at 5000rpm for 10s, place in a water bath at 37°C for 2-4h, and use 1% agarose gel electrophoresis to cut out the target band under UV light , Recovery and purification were carried out with the Gel Recovery Kit. Finally, it was eluted with 30 μl sterile water. Take 2 μl samples for 1% agarose electrophoresis, and store the rest at -20°C for later use. In this step, pEGFP-N1 was cut by Afl II single enzyme.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com