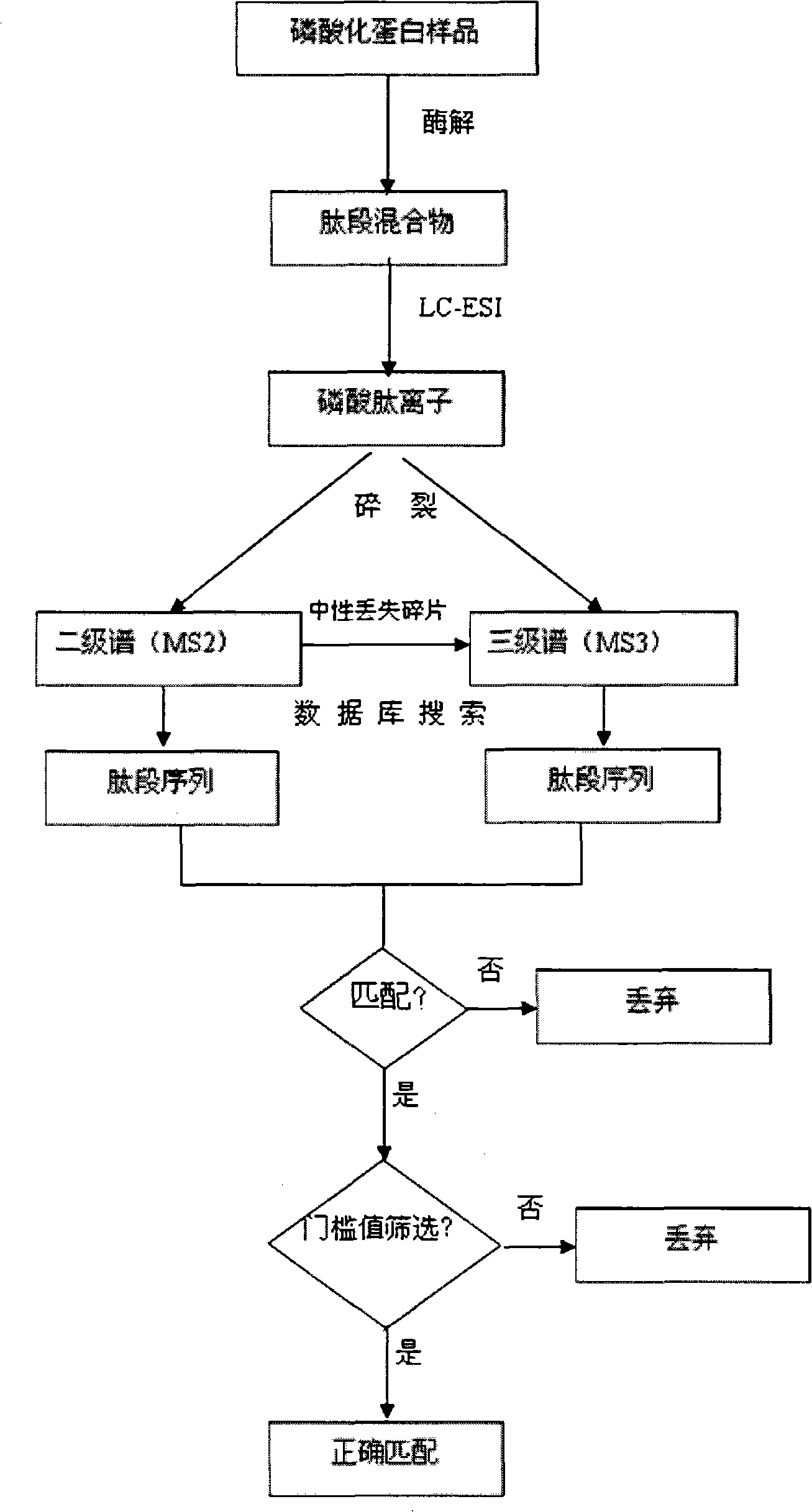
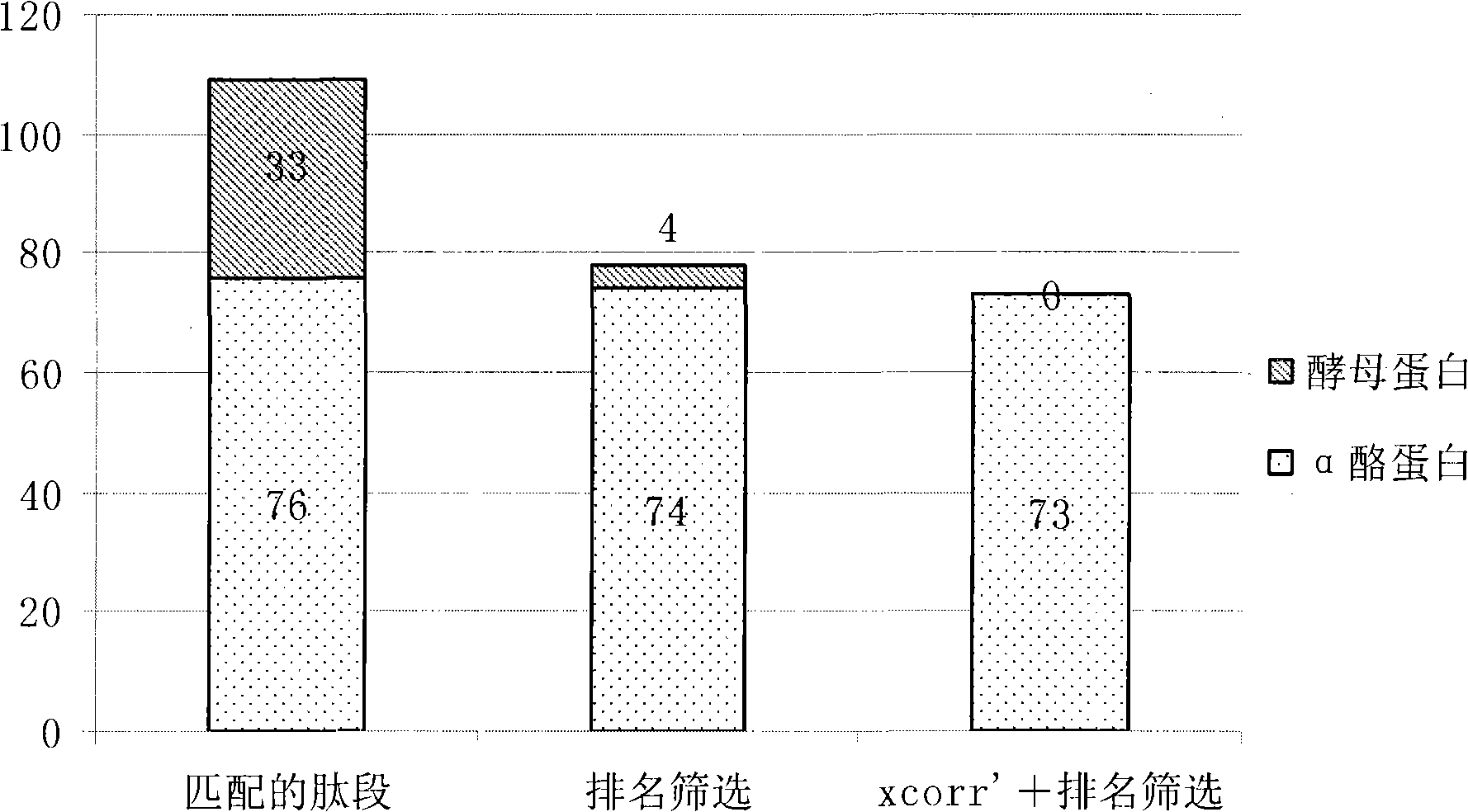
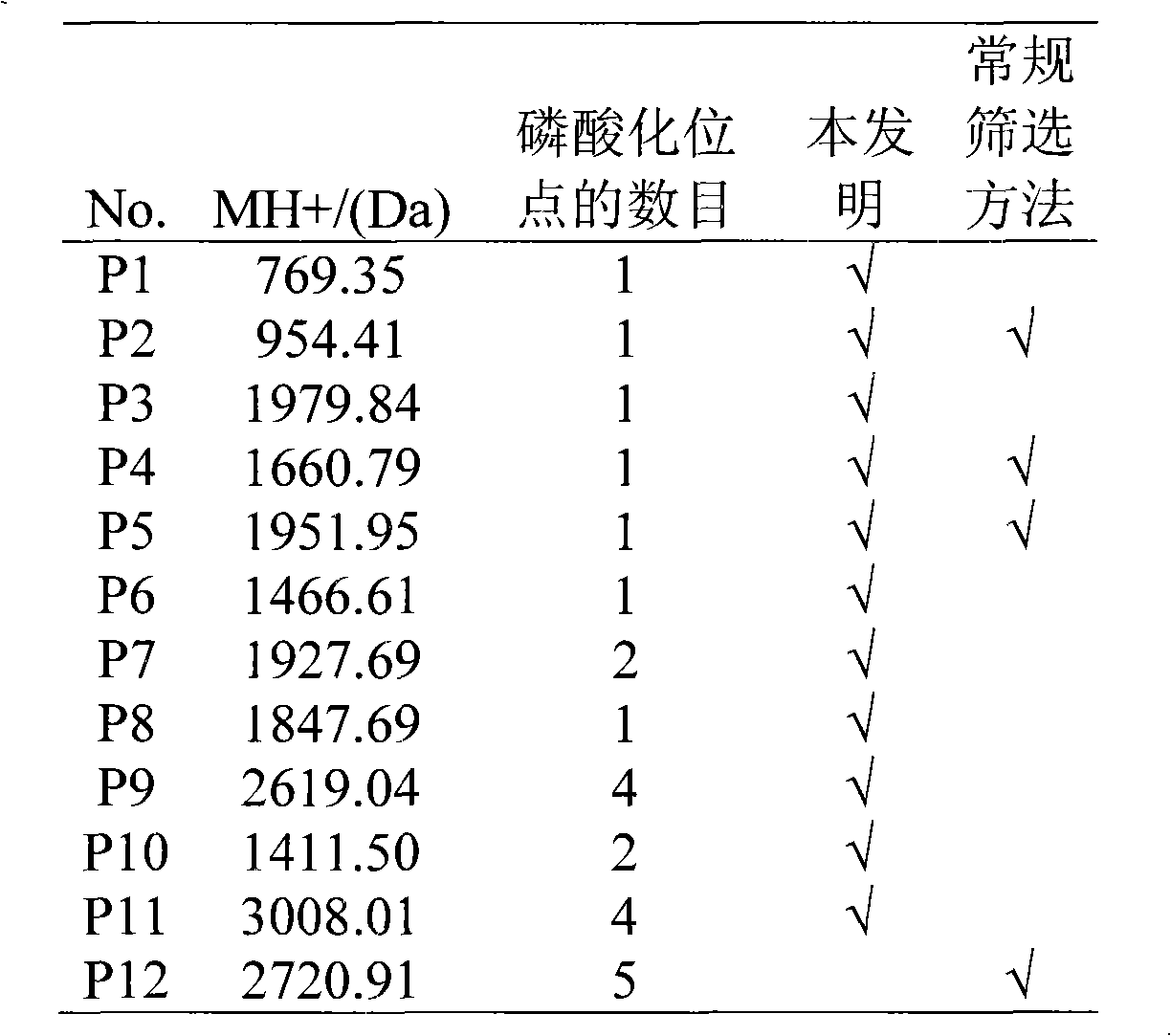
Data processing method for automatically rapid identifying protein phosphorylation site
A data processing and phosphorylation technology, applied in the detection and identification of protein phosphorylation sites, to achieve the effect of improving reliability, avoiding human factors, and high reliability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] The sample preparation and data collection are the same in Example 1 and Example 2, and the specific process is as follows:
[0031] Enzymatic hydrolysis conditions for protein: the buffer is phosphate buffer at pH 8.5, and the concentration of urea is 8 mol / L. After the protein sample was dissolved in the buffer, the disulfide bonds were first reduced with dithiothreitol at 37°C for a reaction time of 2 hours. Then use iodoacetamide to block the sulfhydryl group, and the reaction is to block the light for 2h. Then trypsin was used to digest the reaction at 1:50 overnight at 37°C.
[0032] Mass spectrometry detection conditions for phosphopeptides: The phosphopeptide mixture is analyzed by capillary liquid chromatography-ion trap mass spectrometry, wherein the capillary liquid chromatography adopts a C18 capillary column with an inner diameter of 75 μm, and the mobile phase is (A) 0.1% formic acid aqueous solution (B) 0.1 % formic acid aqueous solution / acetonitrile, b...
Embodiment 2
[0040] Example 2 Applied to the Identification of Phosphorylation Sites in Proteomic Samples
[0041] Extract the liver paracancerous tissue protein from liver cancer patients, and the extracted protein concentration is 2 μg / μL by Coomassie Brilliant Blue Quantitative Method. After 1 mg is digested with trypsin at 37°C for 16 hours, the enzymatic hydrolyzed peptide is directly loaded on the SAX chromatogram For column enrichment, solvent A (40 mM NH4Cl / 30% acetonitrile, pH 4.0) was equilibrated for 5 min, and then eluted by gradient. The elution solvents are solvent A and solvent B (1M NH4Cl / 30% acetonitrile, pH 4.0); the gradient setting is: 0-10min, and the composition of the eluent increases linearly from 100% solvent A to 100% solvent B, and then uses 100% Solvent B eluted isocratically. A fraction was collected every 2 minutes, divided into 8 fractions and collected separately. After the sample was desalted, freeze-dried and reconstituted, 1 / 4 of each fraction was analyz...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com