Zone 2 protein of programmed cell death protein 2 analogue and uses thereof
A technology of programmed death, analogs, applied to the protein fragment of programmed death protein analogs - domain 2 protein field
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] The region 2 protein of the programmed death protein 2 analog is prepared by prokaryotic bacterial fusion expression and then cutting out the amino acid sequence of the carrier, and the preparation method includes the following steps:
[0041] (1) RNA was extracted from gastric cancer cell line SGC-7901 by Trizol method using Trizol kit (purchased from Invitrogen Company), and PrimeScript 1 was used to extract RNA st The strand cDNA Synthesis Kit (purchased from Takara Company) synthesized cDNA as a template for PCR by the method of reverse transcription;
[0042] Use Taq enzyme PrimeSTAR HS DNA Polymerase (purchased from Takara Company) to combine the following primers: ATA GATATC ccagaccacaa ctt cc (GATATC is the EcoRV restriction site) and CCG CTC GAG tta cgg tgt atc tgt tac (CTCGAG is the XhoI restriction site) , pull out the nucleic acid sequence of PDCD2like region 2 by the method of PCR amplification; the PCR conditions are:
[0043] 95°C for 3min, 35cycles of ...
Embodiment 2
[0047] The region 2 protein of the programmed death protein 2 analog is prepared by prokaryotic bacterial fusion expression and then cutting out the amino acid sequence of the carrier, and the preparation method includes the following steps:
[0048] (1) Use the RNeasy mini kit purchased from Qiagen to obtain RNA from the liver cancer cell line HepG2, and use PrimeScript 1 st The strand cDNA Synthesis Kit (purchased from Takara Company) synthesized cDNA as a template for PCR by the method of reverse transcription;
[0049] Use Taq enzyme PrimeSTAR HS DNA Polymerase (purchased from Takara Company) to combine the following primers: ATA GATATC ccagaccacaa ctt cc (GATATC is the EcoRV restriction site) and CCG CTC GAG tta cgg tgt atc tgt tac (CTCGAG is the XhoI restriction site) , pull out the nucleic acid sequence of PDCD2like region 2 by the method of PCR amplification; the PCR conditions are:
[0050] 95°C for 3min, 35cycles of 98°C for 15s, 45°C for 30s, 72°C for 1min, the PC...
Embodiment 3
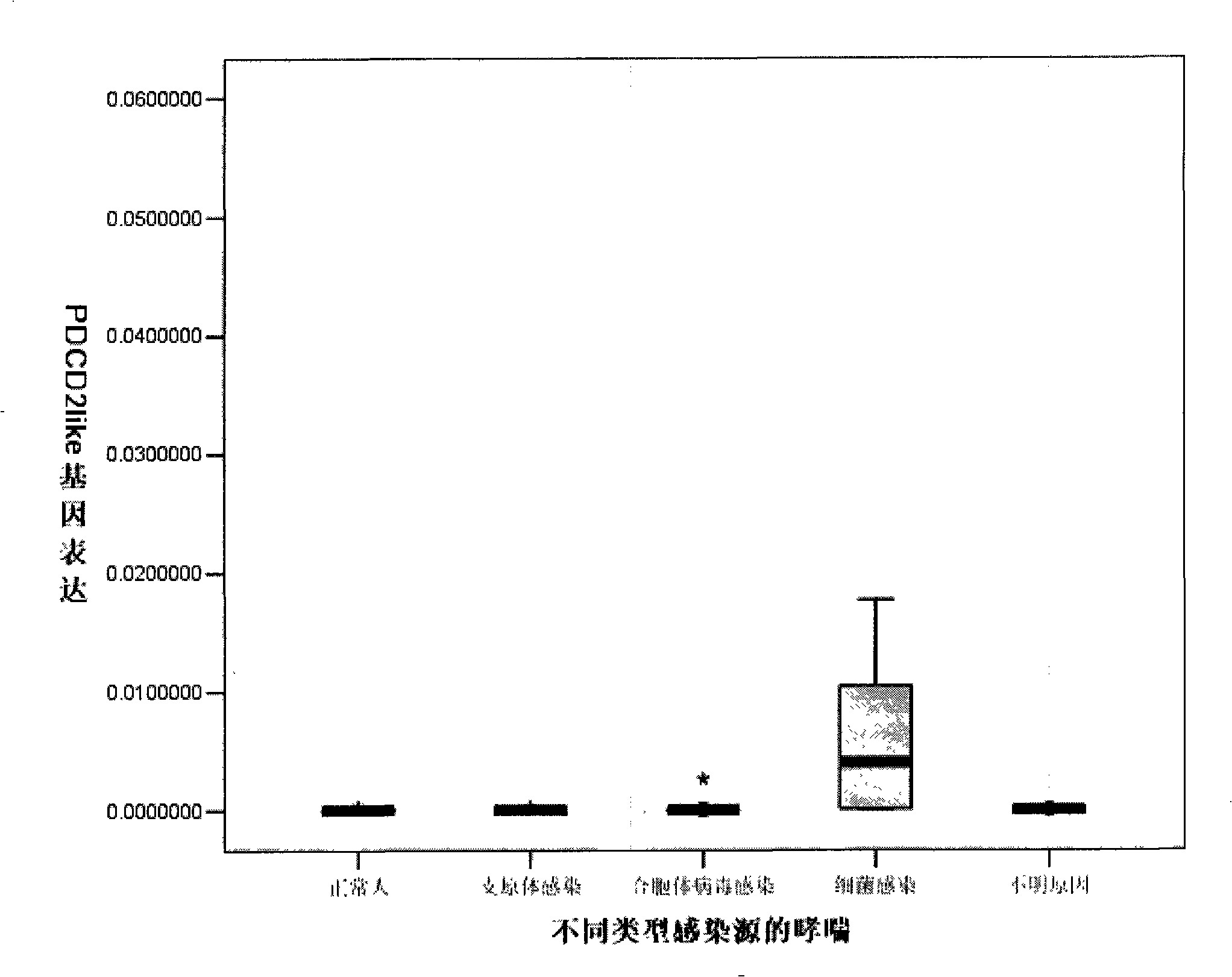
[0054] 98 children with asthma: 35 cases of syncytial virus infection, 19 cases of bacterial (Streptococcus pneumoniae, Moraxella catarrhalis, influenza (Haemophilus) bacillus) infection, 9 cases of mycoplasma infection, 35 cases of unknown etiology, 13 cases of normal children , extract 1 ml of peripheral blood, separate white blood cells with ficoll separation medium, add 1 ml Trizol to extract RNA, use reverse transcriptase to reverse transcribe the RNA into cDNA, and analyze the expression of programmed death protein 2 analog domain 2 by real-time quantitative PCR 18s rRNA was used as internal reference. Base 2, C of 18s rRNA T Value minus programmed death protein 2 analogue region 2 C T The value is an exponent, ie: 2 -(CT,PDCD2like-CT,18s rRNA) , represents the relative expression level of programmed death protein 2 analog domain 2 in each sample.
[0055] from image 3 It can be seen that the expression of programmed death protein 2 analog domain 2 in patients with ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com