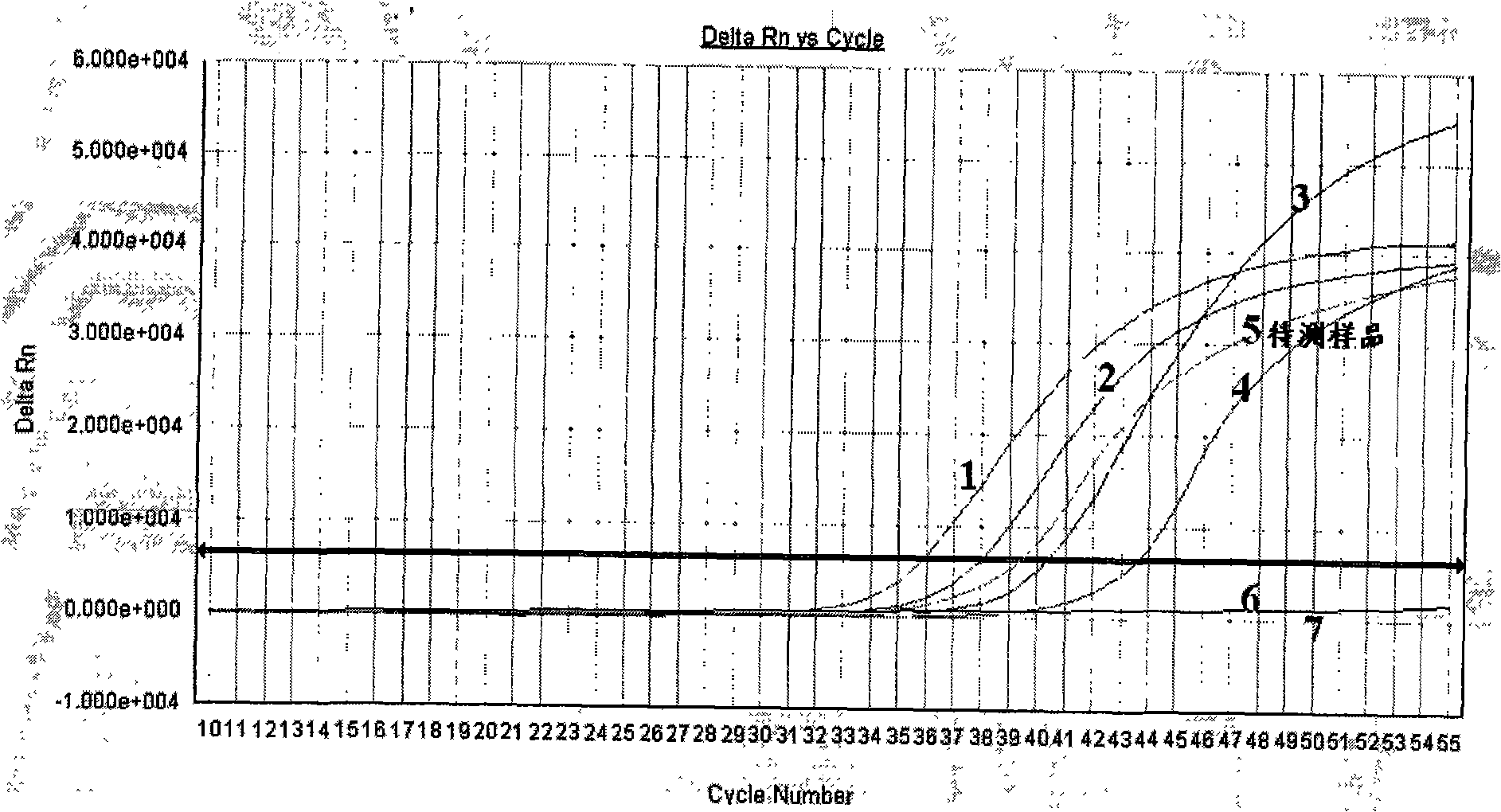
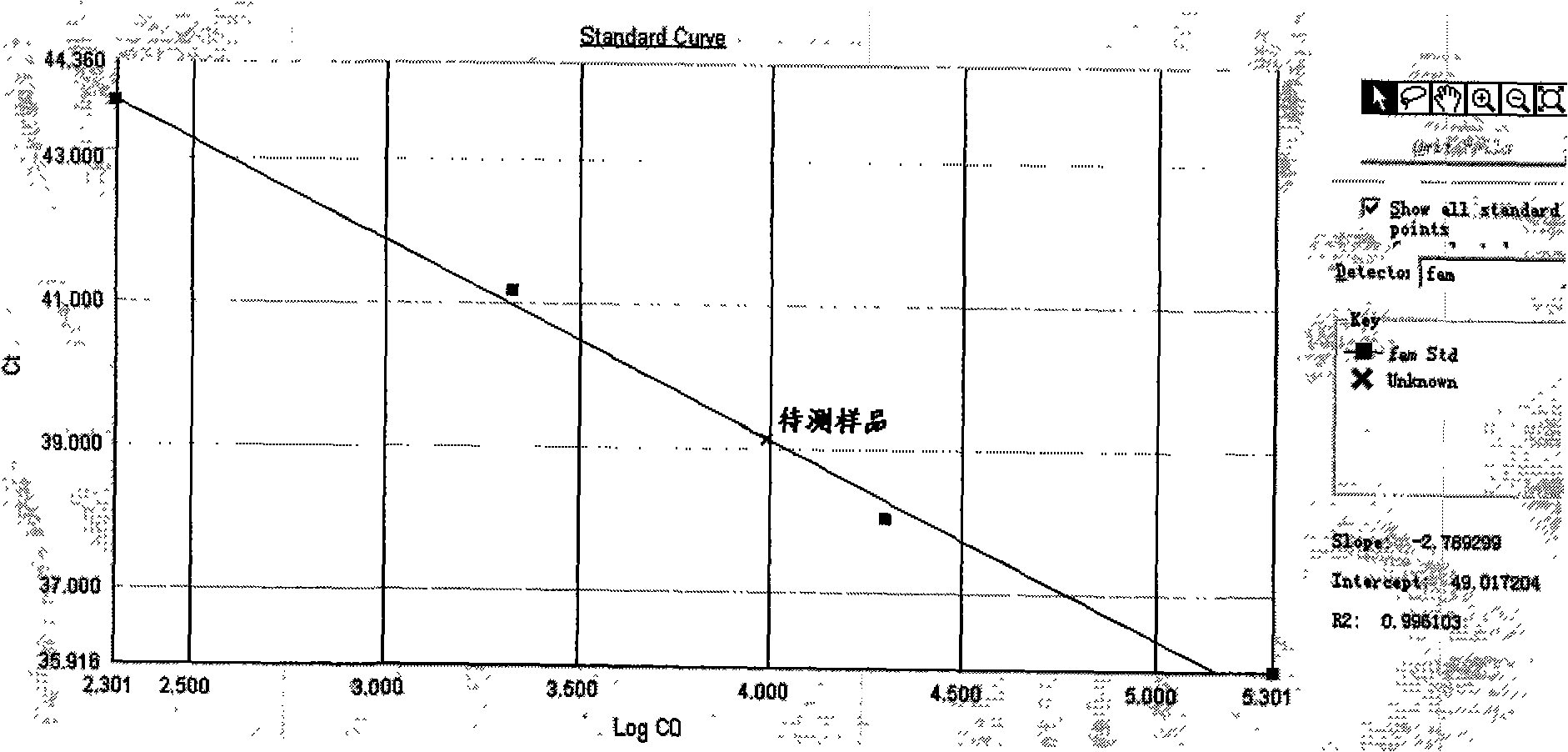
Methylation quantitative detection method of RASSFIA gene in human plasma
A technology of methylation quantification and detection method, applied in the field of biomacromolecule detection, can solve the problems of environmental pollution, missed diagnosis, low sensitivity and accuracy, etc., and achieve the effect of strong promotion, less trauma and strong specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0037] 1. Plasma Separation and Storage
[0038] with EDTA-K 2Extract 2ml of human peripheral venous blood from a 5ml sterile plastic anticoagulant tube with a cover, and centrifuge at 3000rpm for 10min at room temperature for 48 hours to collect plasma; centrifuge again at 12000rpm for 10min to obtain blood cell-free plasma The tubes are divided into 200 μl tubes, stored at a temperature below -20°C, and tested within 1 month.
[0039] Note: Glass centrifuge tubes cannot be used to collect anticoagulated venous blood.
[0040] 2. Preparation of plasma DNA standard curve samples
[0041] (1) Lymphocytes were separated from healthy fetal cord blood (CB) by Ficoll lymphocyte separation medium, and CB DNA was extracted by traditional phenol / chloroform method;
[0042] (2) CB DNA transmethylation: 10×NEB buffer 2μl, 1.6mM SAM 2μl, SssI Methylase 4U, CB DNA 1μg, total volume 20μl, positive control DNA was prepared according to standard operating procedures;
[0043] (3) Prepara...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com