Host cell and method for efficient expression and secretion thereof in recombinant protein
A host cell, secretion and expression technology, which is applied in the field of microbiology and fermentation engineering, can solve the problems of high efficiency, secretion and expression, induction, and long culture time of recombinant proteins, so as to reduce the environmental pressure of fermentation industry, reduce the cost of fermentation manufacturing, The effect of simplifying the fermentation manufacturing process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0071] Example 1: Screening of Bacillus licheniformis host cells
[0072] The selection process of Bacillus licheniformis host cells is shown in the figure below (attached figure 1 ).
[0073] 526 strains of Bacillus licheniformis were isolated from 3050 natural samples collected in China, and screened against Bacillus licheniformis ATCC14580 and CICIM B0204. All collected Bacillus licheniformis strains were inoculated with LB medium supplemented with 4% glucose, cultured at 45°C for 72 hours, observed the color of the colonies and the formation of pigments around the colonies, and determined the formation of lichenin and polyglutamic acid by staining. 24 strains of Bacillus licheniformis were selected for the next test. 24 strains were inoculated on 0.01 mg / mL tetracycline or 0.005 mg / mL kanamycin resistance plates respectively, and after culturing at 37°C for 24 hours, 18 of them were determined to be antibiotic-sensitive. The possible self-plasmids were extracted and ide...
Embodiment 2
[0078] Example 2: Cloning of restriction modification system encoding gene cluster and construction of gene deletion plasmid
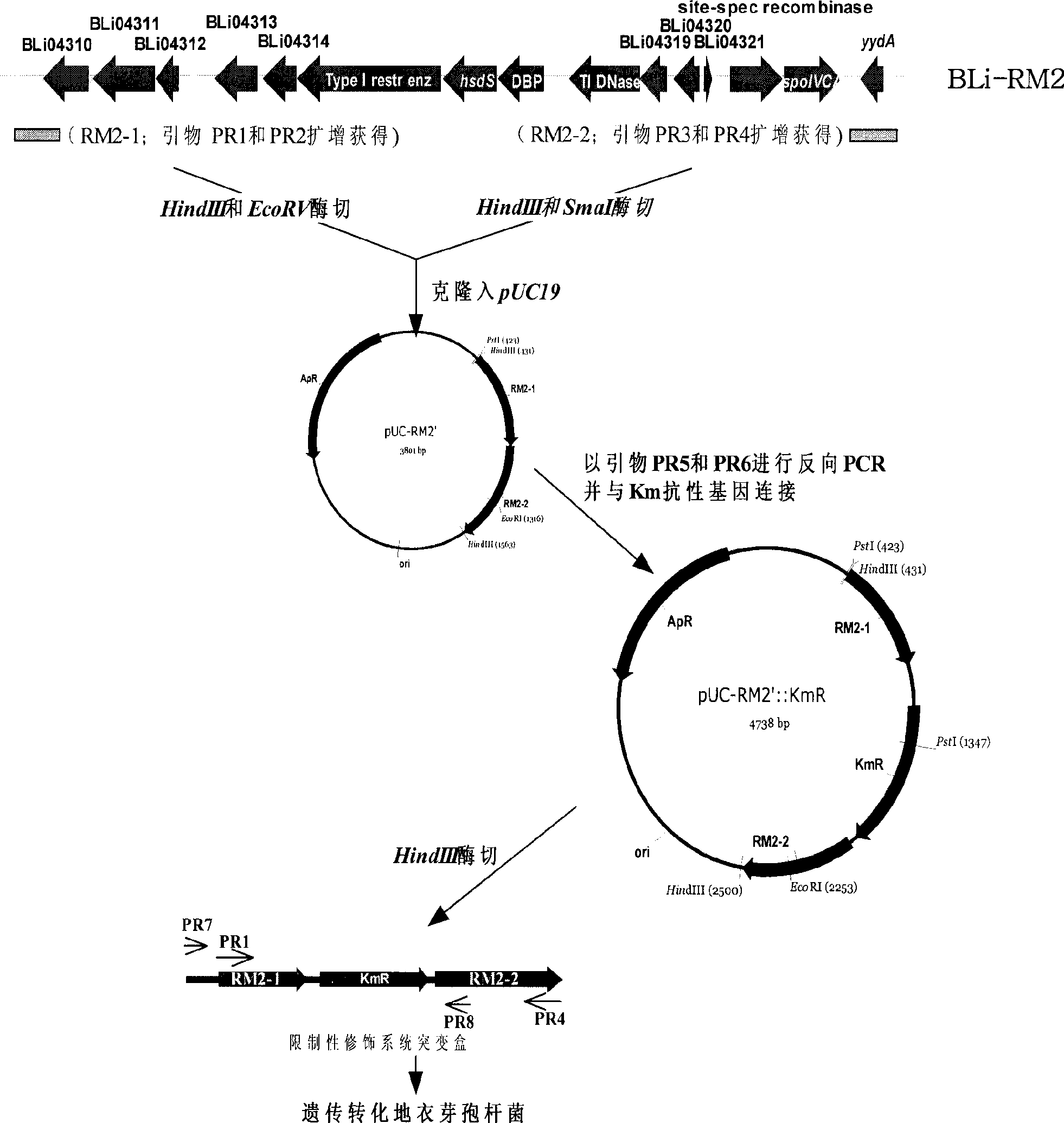
[0079] Analysis of the genome sequence of B. licheniformis ATCC 14580 shows that in the region of about 17.3 kb from 4157995 bp to 4175263 bp of the genome, there is a B. licheniformis specific type I DNA restriction modification system coding gene cluster (Veith B, et al. J Mol Biotechnol, 2004). Design the following steps to delete it (attached image 3 ).
[0080] Using primers PR1 (SEQ ID NO: 1) and PR2 (SEQ ID NO: 2), PCR technology was used to amplify the upstream sequence RM2-1 of the restriction modification system gene cluster from the chromosomal DNA of Bacillus licheniformis 3008, with a size of about 560bp, ( Figure 4A); Similarly, using primers PR3 (SEQ ID NO: 3) and PR4 (SEQ ID NO: 4), the downstream sequence RM2- 2, the size is about 610bp ( Figure 4 A). Fragment RM2-1 was digested with HindIII and EcoRV and fragment RM2-2 was di...
Embodiment 3
[0081] Example 3: Deletion of DNA Restriction Modification System Encoding Gene Clusters
[0082] The recombinant plasmid pUC-RM2'::Km R Amplification was carried out in recombinant bacteria Ec(met) (Wang Zhengxiang, Niu Dandan. Chinese invention patent, publication number: CN 101230329A). After the preparation of the recombinant plasmid was digested with HindIII, the Bacillus licheniformis 3008 cells were transformed by electroporation. The transformants were spread on LB plates containing 0.025 mg / mL kanamycin and cultured for 48 hours. After the growth colony was subcultured and purified on the LB medium of 0.025 mg / mL kanamycin, it was then subcultured in the LB medium without kanamycin, and the bacterial strain that kanamycin resistance disappeared was screened, and the primer PR7( SEQ ID NO: 7) and PR8 (SEQ ID NO: 8) identify kanamycin-sensitive strains, and the restriction modification system coding gene cluster in Bacillus licheniformis 3008 is deleted, then primer P...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com