Fluorescent detection kit of cholera vibrio O139 and detection method
A technology for Vibrio cholerae and fluorescence detection, which is applied in the direction of fluorescence/phosphorescence, microbial measurement/inspection, biochemical equipment and methods, etc. It can solve the problems of inaccurate detection results, false positives, long cycle and so on, so as to reduce false positives Rate, Sensitivity Reduction, Sensitivity Improvement Effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
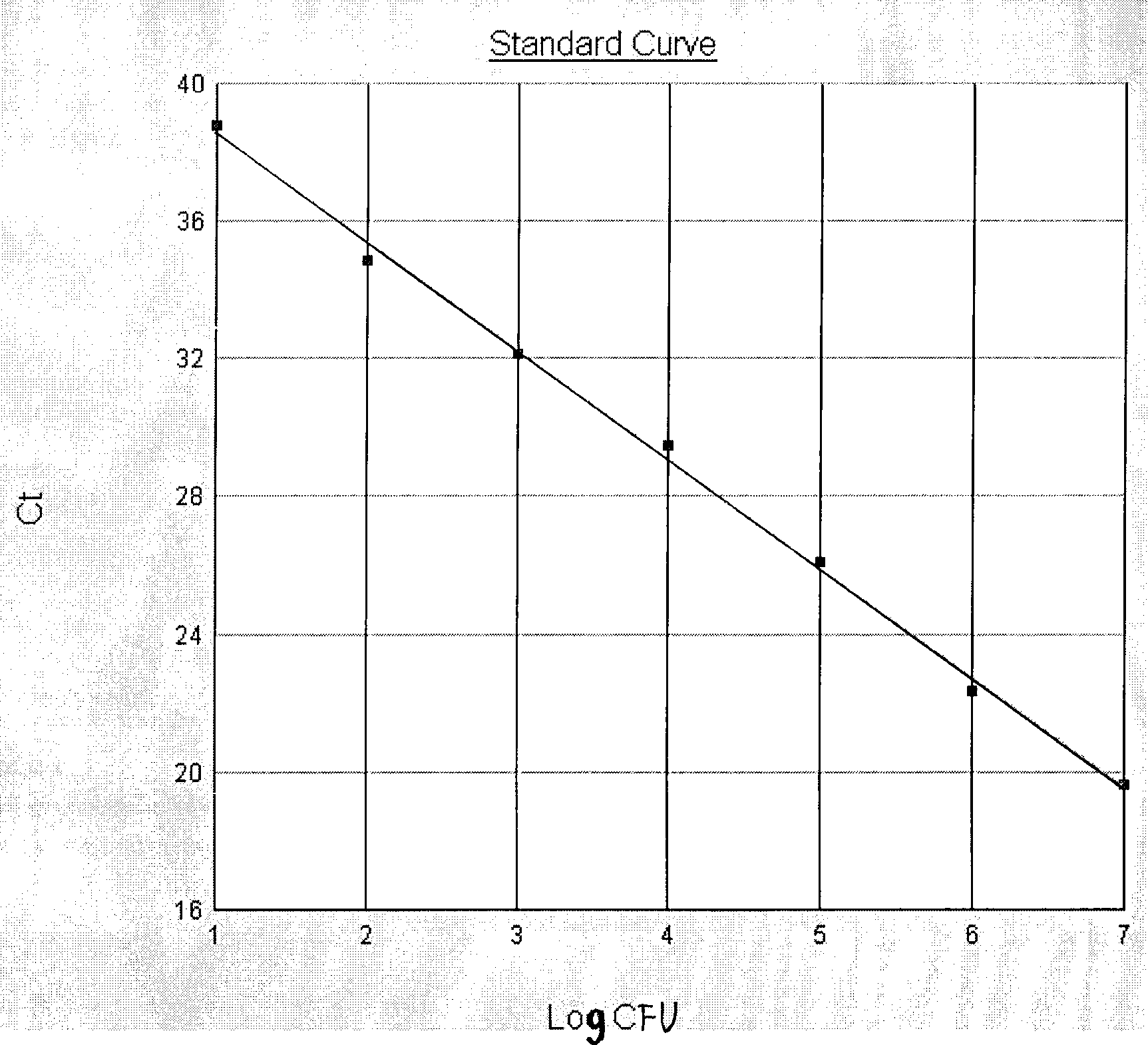
Image
Examples
Embodiment 1
[0030] Embodiment 1: the acquisition of specific primers, fluorescent probes and standards
[0031] 1. Materials:
[0032]Bacterial genomic DNA extraction reagents were purchased from Dalian Bao Biological Engineering Co., Ltd.; restriction endonucleases were purchased from LTI / Gibco, USA; pGEM-T-Easy cloning system, PCR buffer, and TaqDNA polymerase were purchased from Promega, USA; sequencing reagents, Model 377 sequencer, Bio-Radicycler PCR instrument and Model 7000 quantitative PCR instrument are all products of American ABI Company.
[0033] 2. Primer and probe design and synthesis:
[0034] Using the Vibrio cholerae O139 glycosyltransferase gene sequence (registration number U72485) as a template, use Primer Express TM (V2.0, American ABI Company) software analyzes TaqMan primer and probe site, selects the optimal combination therefrom.
[0035] Standard PCR upstream primer sequence is:
[0036] 5'-TGTAATGATT AGAAAGAAAG TTA-3',
[0037] Downstream primers are:
[0...
Embodiment 2
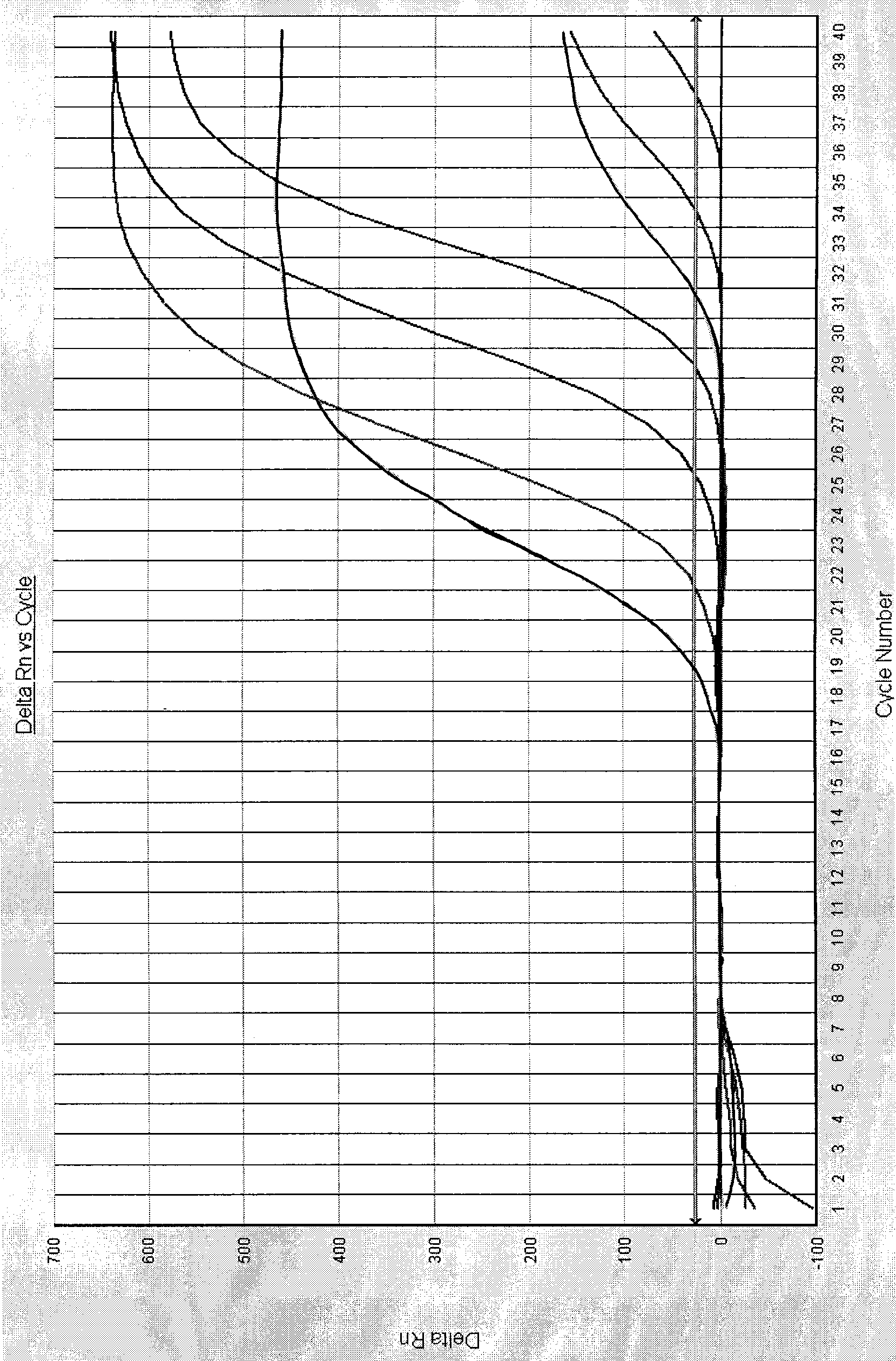
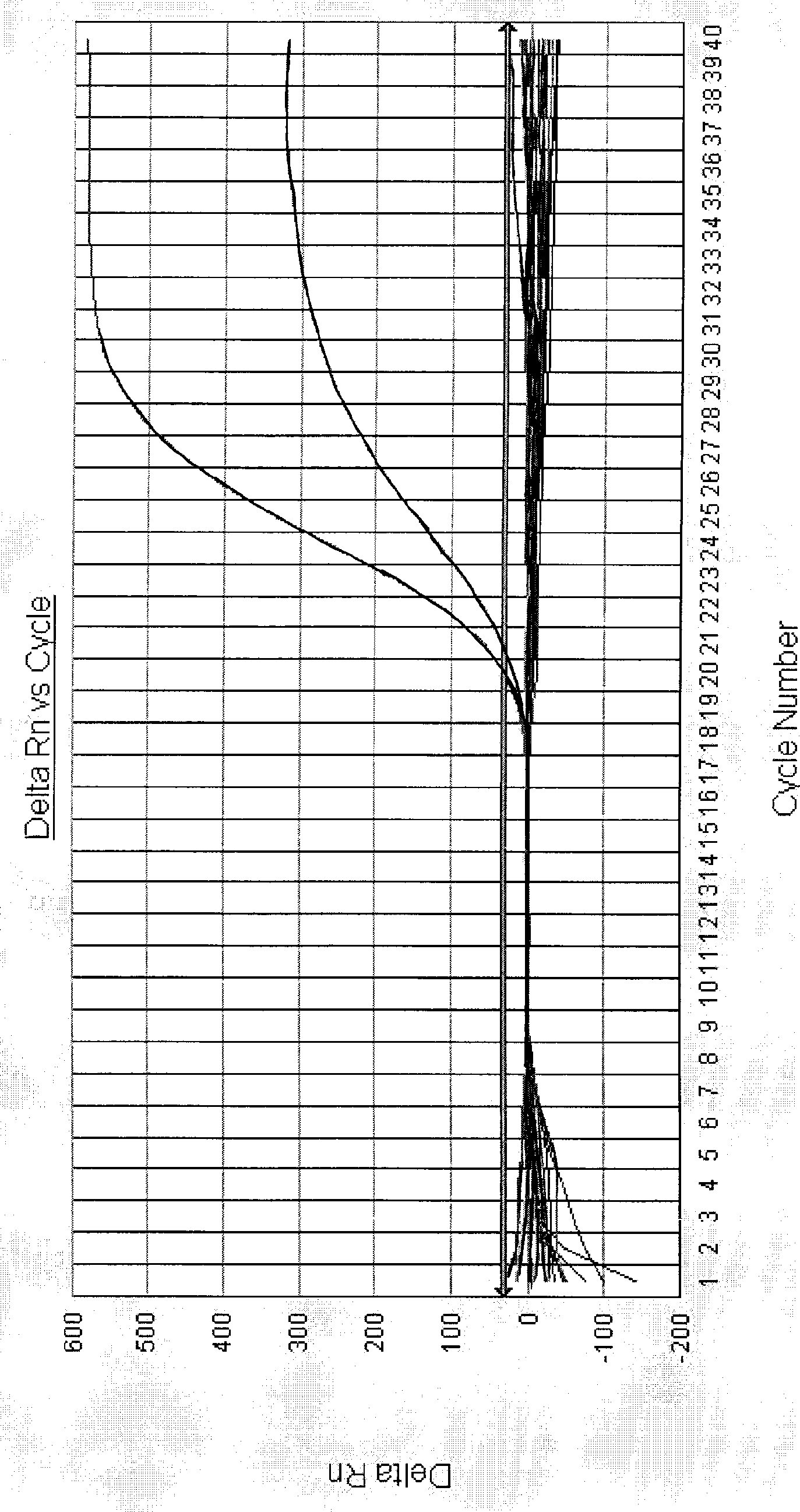
[0061] Embodiment 2: Fluorescent quantitative PCR method detects Vibrio cholerae O139
[0062] 1. Specimen detection:
[0063] 52 cases of clinical stool samples were washed with normal saline and then centrifuged. Genomic DNA was extracted with genomic DNA extraction reagents. 1.0 μL was taken as a template, and PCR amplification was carried out on a 7000 quantitative PCR instrument of ABI Company with upstream and downstream primers for detection.
[0064] The composition of the PCR reaction solution is as follows:
[0065] 2×PCR buffer 10.0μL
[0066] Detection upstream primer (10μM) 1μL
[0067] Downstream primer for detection (10μM) 1μL
[0068] Fluorescent probe for detection (10μM) 1μL
[0069] Taq DNA polymerase (5U / μL) 0.2μL
[0070] dNTPs (250mM each) 1.60μL
[0071] (dATP, dTTP, dCTP, dGTP substance ratio 1:1:1:1)
[0072] Template DNA (50ng / μL) 1μL
[0073] Make up to 20 μL with water.
[0074] The PCR reaction conditions were: pre-denaturation at 93°C for...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com