Uracil auxotroph Hansenula yeast, construction method thereof and application thereof
A technology of auxotrophic and Hansenula spp., applied in the direction of microorganism-based methods, biochemical equipment and methods, enzymes, etc., can solve the problem of low expression level of exogenous genes, low level of expressed protein secretion, low level of polypeptide chain sugar Over-transformation and other problems can be achieved to achieve the effect of low reverse mutation rate, short culture time and fast growth rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
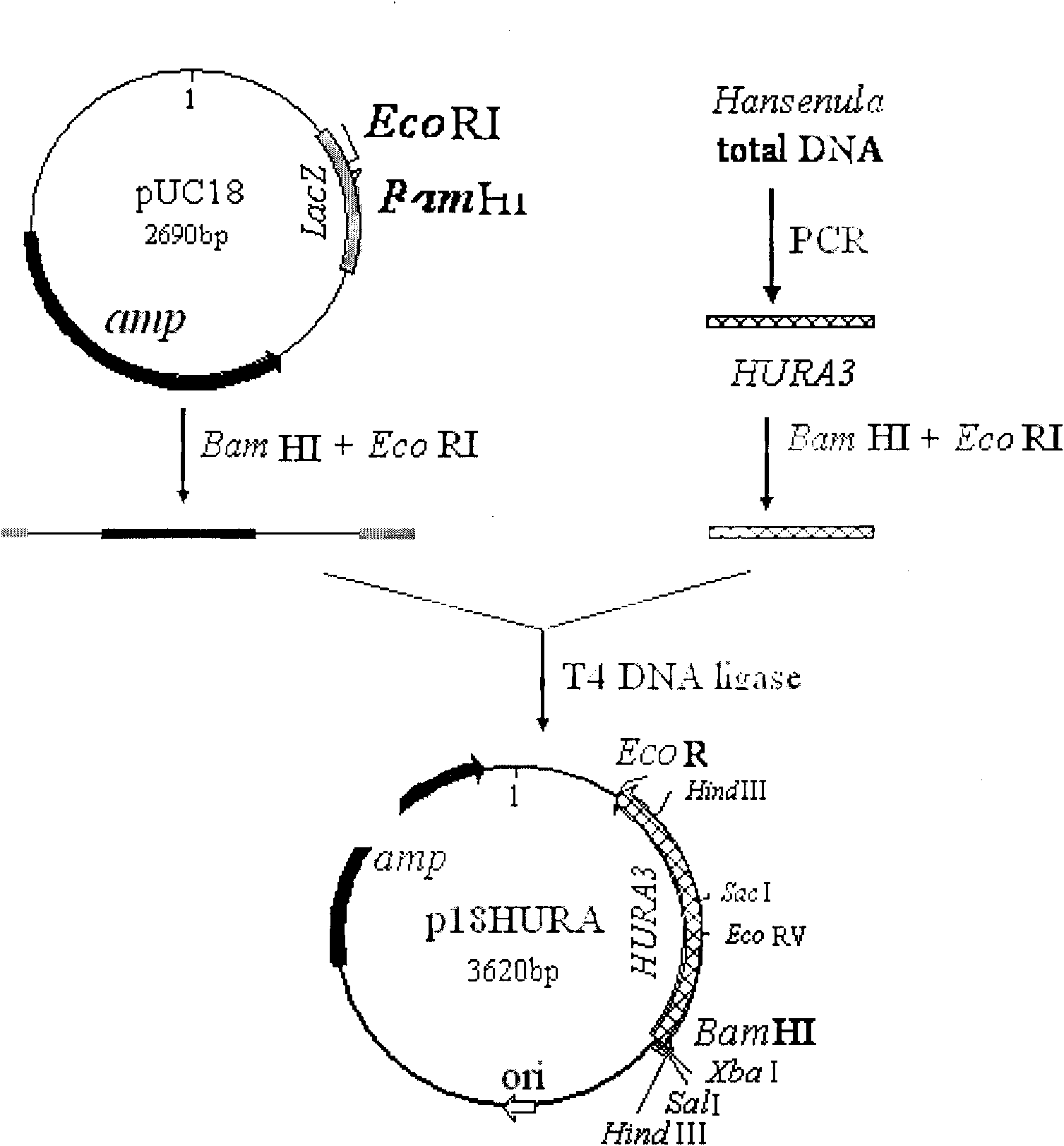
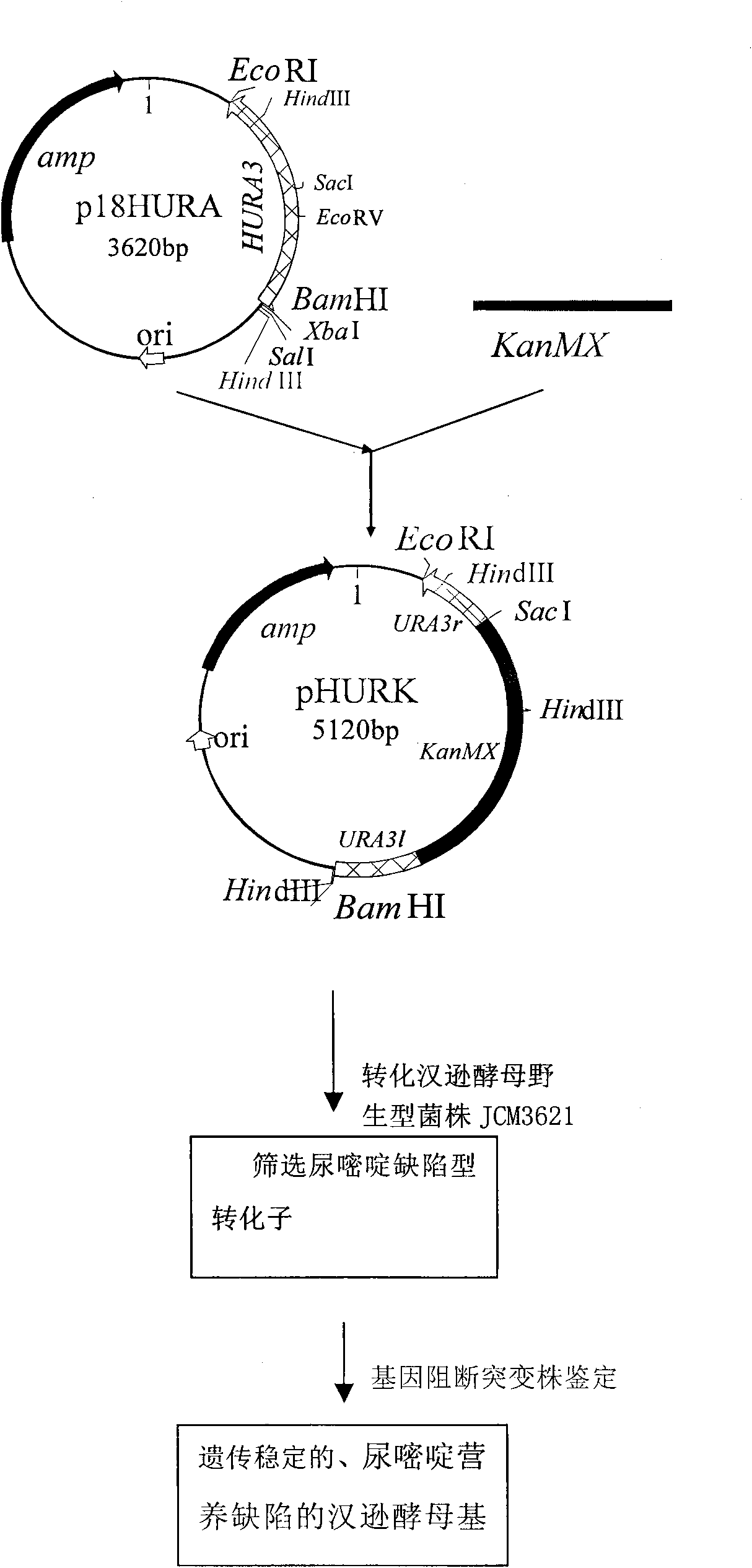
[0036] Construction of Hansenula
[0037] 1. Construction of the recombinant plasmid p18HURA containing the orotidine-5-phosphate decarboxylase gene (HURA3) of Hansenula spp. The construction process of the recombinant plasmid p18HURA is as follows:
[0038] 1. Acquisition of orotic acid-5-phosphate decarboxylase gene (HURA3) of Hansenula spp.
[0039] The primer sequences designed according to the reported nucleotide sequence of Hansenula HURA3 are as follows:
[0040] Primer 1: 5′-CCA GGATCC TCAACATTTCCCTGAATAAT-3' (sequence 1 in the sequence listing) (the bases underlined are BamH I recognition sites)
[0041] Primer 2: 5′-CGA GAATTC TCACTAGTATTC CCGCGACT-3' (sequence 2 in the sequence listing) (the underlined part of the base is the EcoR I recognition site)
[0042] Using the total DNA of Hansenula (Hansenula) JCM3621 (ATCC34438) as a template, PCR reaction was carried out to amplify HURA3 under the guidance of primers 1 and 2. The PCR reaction system was: template D...
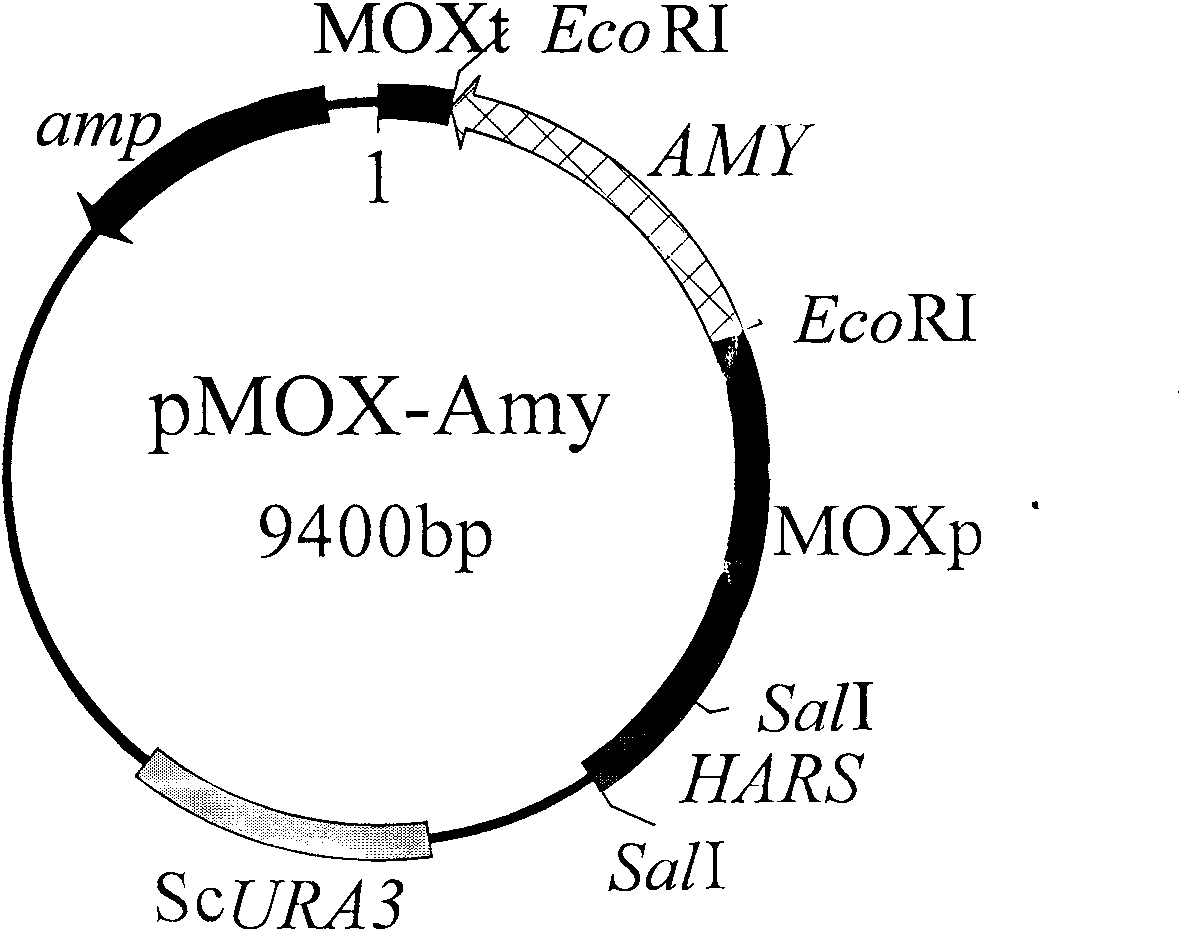
Embodiment 2
[0064] Detection of biomass and biological activity of Hansenula YH-11CGMCCNO.2976, a uracil-auxotrophic Hansenula genetically engineered host strain:
[0065] Detection of Biomass of Uracil Auxotroph Hansenula YH-11
[0066] Inoculate wild-type Hansenula and uracil-auxotrophic Hansenula YH-11 on YEPD slant medium for activation, pick a ring of bacteria from the slant with an inoculation loop and inoculate 2ml of YEPD In liquid culture medium, shake culture at 37°C for 16 hours; transfer to 10ml YEPD liquid medium according to 10% inoculum size, shake culture at 37°C for 16 hours; collect bacterial cells by centrifugation, and wash the cells with sterile water for two Second, after weighing, the cells are divided into four equal parts, two parts of cells are one group, two groups of cells are respectively inoculated in 50ml YEPD liquid medium and 50ml in the liquid medium of glucose in YEPD with methanol, Then two copies of cells in each group were shaken and cultured at 37°C...
Embodiment 3
[0068] Genetic stability analysis of uracil-auxotrophic Hansenula genetically engineered host strain Hansenula YH-11CGMCCNO.2976.
[0069] Genetic stability analysis of uracil auxotroph Hansenula YH-11
[0070] Inoculate the uracil auxotrophic Hansenula YH-11 constructed above with blocked HURA3 gene function in YEPD liquid medium and subculture for 30 times, take 200 μl of diluted cells and spread them on the YEPD plate, Incubate at 37°C for 48 hours, randomly pick 100 single colonies and place them in 2ml sterile water, and starve them for 4-6 hours at room temperature; take the starved bacteria solution and inoculate them on YNB plates with glucose as the carbon source and add 35μg / ml uracil on the YNB plate, and on the YNB plate using methanol as a carbon source and the YNB plate adding 35 μg / ml uracil, cultured at 37°C for 48 hours, the results showed that single colonies were all on the YNB basic medium added uracil. growth, on the YNB basic medium without adding uracil,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com