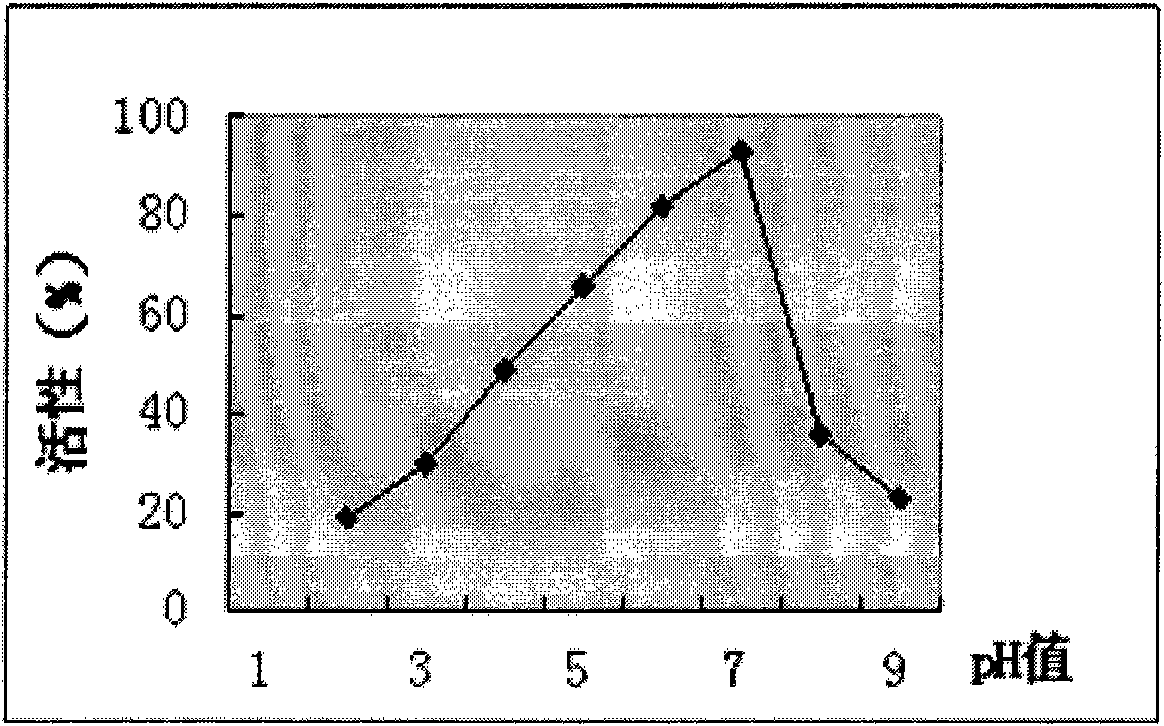
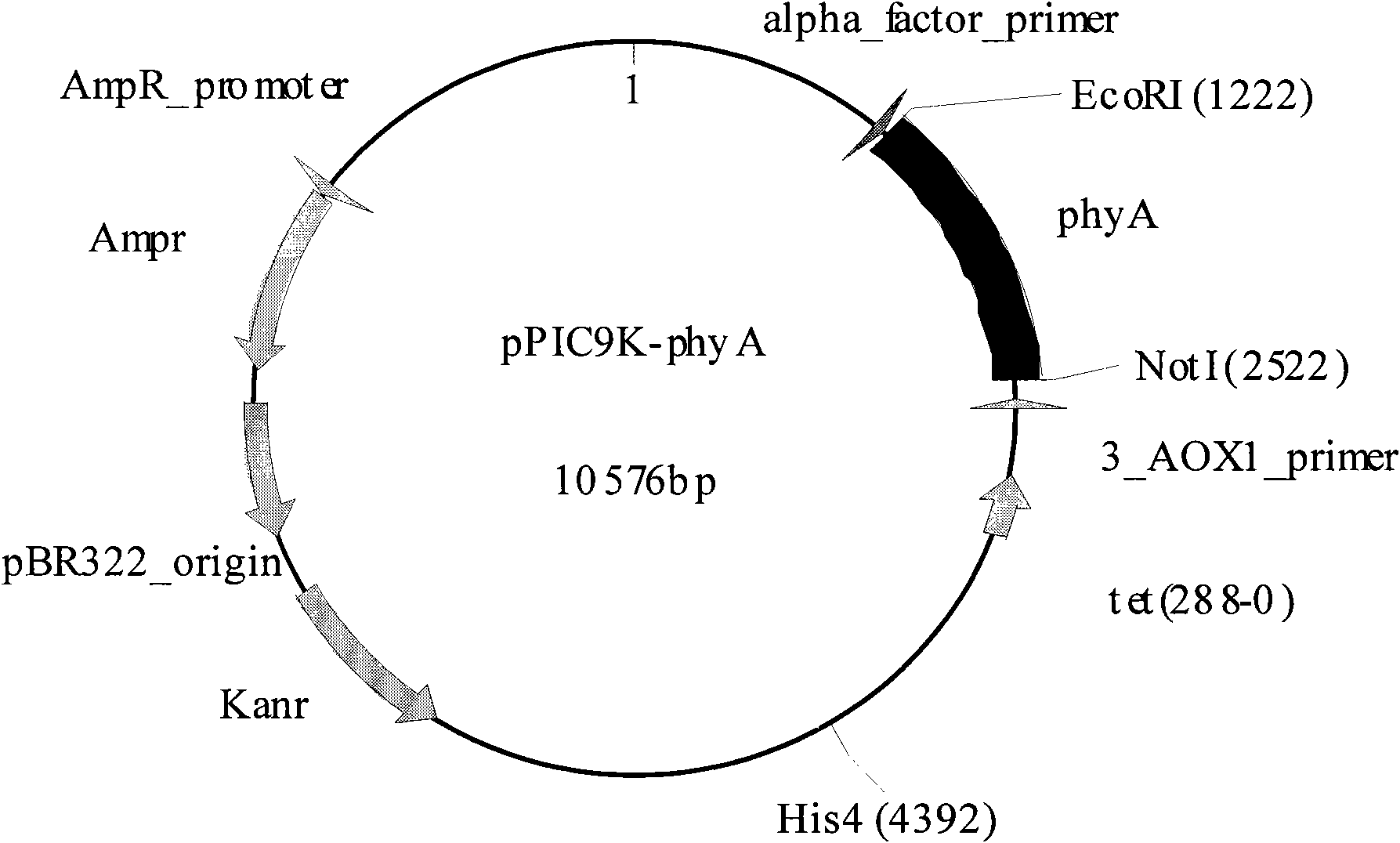
Novel neutral phytase
A technology of phytase and phytase protein, applied in hydrolytic enzymes, plant genetic improvement, botany equipment and methods, etc., can solve problems such as difficult extraction production and low phytase content
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Embodiment 1 produces the screening of the Citrobacter of phytase
[0028] Soil was obtained from high-temperature irradiation environment in Xinjiang for microbial enrichment culture. The medium uses LB medium, and its ingredients are: 0.5% yeast powder, 0.3% peptone, 0.5% sodium chloride, and pH 7. Culture conditions: Weigh an appropriate amount of soil and put it into LB medium, 37°C, 200r / min, and cultivate for 3-6 days. Draw a small amount of culture solution every day for gradient dilution (10-2-10-10), spread it on a solid medium containing sodium phytate, and culture it at 37°C for 2-3 days. Solid medium composition: sodium phytate 1%, NaNO30.3%, MgSO4·7H2O 0.5%, KCl 0.5%, K2HPO40.1%, FeSO4·7H2O 0.001%, agar 1%, pH6.0. If there are obvious bacterial circles around the colonies grown in the medium, it can be preliminarily judged that the bacteria can produce phytase, and the colonies that produce the largest bacterial circles in the medium can be selected.
Embodiment 2
[0029] Example 2 Extraction and Identification of Citrobacter sp. Genome
[0030] Pick a single colony of the bacteria in the selected medium and put it into LB liquid medium for culture, and take the bacterial liquid for the extraction of the genomic DNA of the bacteria. Using the TIANamp Bacteria DNA Kit Bacterial Genomic DNA Extraction Kit from Tiangen Company, the genomic DNA of the bacteria was extracted according to the operating instructions of the kit provided by the company. In order to identify the genus, the 16s rDNA sequence of the strain was amplified by polymerase chain reaction (PCR) using genomic DNA as a template.
[0031] Design of primers for bacterial 16S rDNA: F 27:5′-agagtttgatcatggctcag-3′
[0032] R 1492: 5′-tacggttacccttgttacgactt-3′
[0033] The length of the PCR product is about 1.4kb. After sequencing, it is compared with the DNA sequence in the GenBank database. It is found that the sequence is 98% similar to the 16s rDNA g...
Embodiment 3
[0034] Cloning of Phytase Gene in Example 3 Citrobacter Genome
[0035] Find the phytase gene sequence in other Citrobacter from Genbank, and design degenerate primers, as follows:
[0036] phyA F(EcoR I): 5′- GAATTC ATGAGTACATTCATCATTC-3′
[0037] phyA R(Not I): 5′- GCGGCCGC TTATTCCGTAACTGCACAC-3′
[0038] The underlined part is EcoRI (upstream primer F) and NotI (downstream primer R) recognition sites.
[0039] After the PCR product was subjected to 0.8% agarose gel electrophoresis, the observed length was about 1.3Kb. T4 DNA ligase was used to connect to the T-vector (pMD18-T simple vector, Takara Company). After sequencing, the deduced amino acid sequence is shown as SEQ ID NO:2.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com