Method for screening medicament taking focal adhesion kinase (FAK) transcriptional regulation as target spot and application
A technology for transcriptional regulation and screening methods, applied in biochemical equipment and methods, microbial assay/inspection, etc., can solve the problems of lack of major breakthroughs in carrier and delivery, and the inability of RNA interference therapy to be practically applied, and achieve the effect of convenient use.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
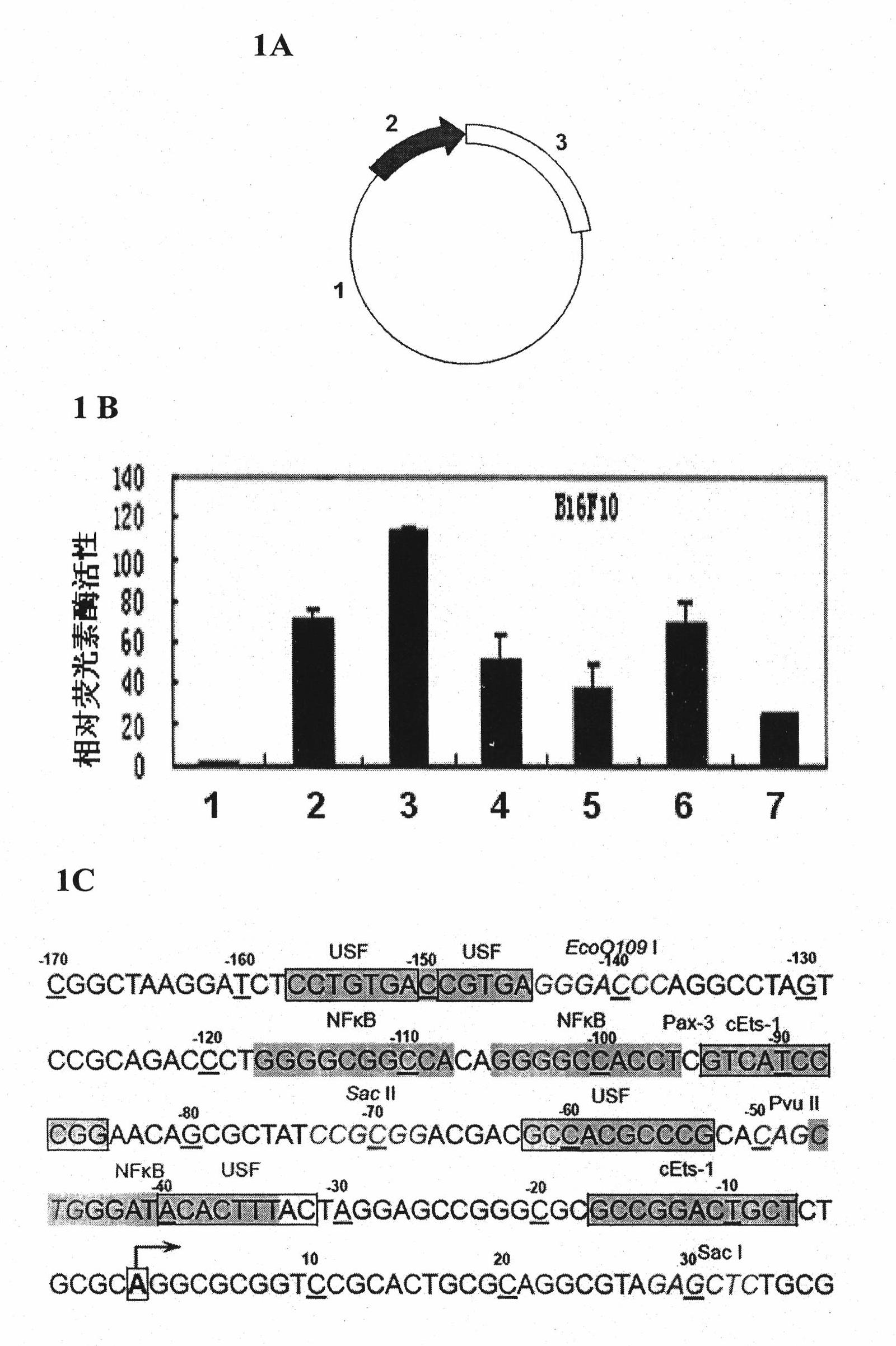
[0072] 1. Construction of mouse FAK promoter reporter gene vector:
[0073] Using the TRANSFAC database and bioinformatics methods to analyze and predict the mouse FAK gene promoter and its transcription factor binding sites, on this basis the mouse FAK promoter and its core sequence were obtained. Design corresponding PCR primers, chemically synthesize upstream and downstream primers (upstream primer: 5'-CGGCTAAGGATCTCCTGTGACCGTGA-3'; downstream primer: 5'-CAGCGCAGAGCTCTACGCCTGCGCAG-3', use the extracted mouse genome as a template, and use the PCR method to amplify The mouse FAK promoter sequence was obtained. The conditions of the PCR reaction were: heat denaturation at 94°C for 1 minute, annealing at 60°C for 1 minute, extension at 72°C for 1 minute, and a total of 25 cycles. The PCR product was recovered by agarose gel electrophoresis Kpn I / Sma I double digestion was carried out, and the reporter gene carrier pGL3-Basic was also subjected to Kpn I / Smal I double digestion. ...
Embodiment 2
[0127] 1. Human genomic DNA extraction:
[0128] Human pancreatic cancer cells SW1990 in the logarithmic growth phase were digested and divided into 35mm culture dishes, and cultured overnight in a cell culture incubator at 37°C and 5% CO2, until the cell density grew to about 80-90%, washed with PBS for 2 The second time, the cells were collected with a cell scraper, and the cell pellet was collected by centrifugation. The genome was extracted according to the instructions of the genome extraction kit. The extracted genome was dissolved in ultrapure water and stored at -20°C for later use.
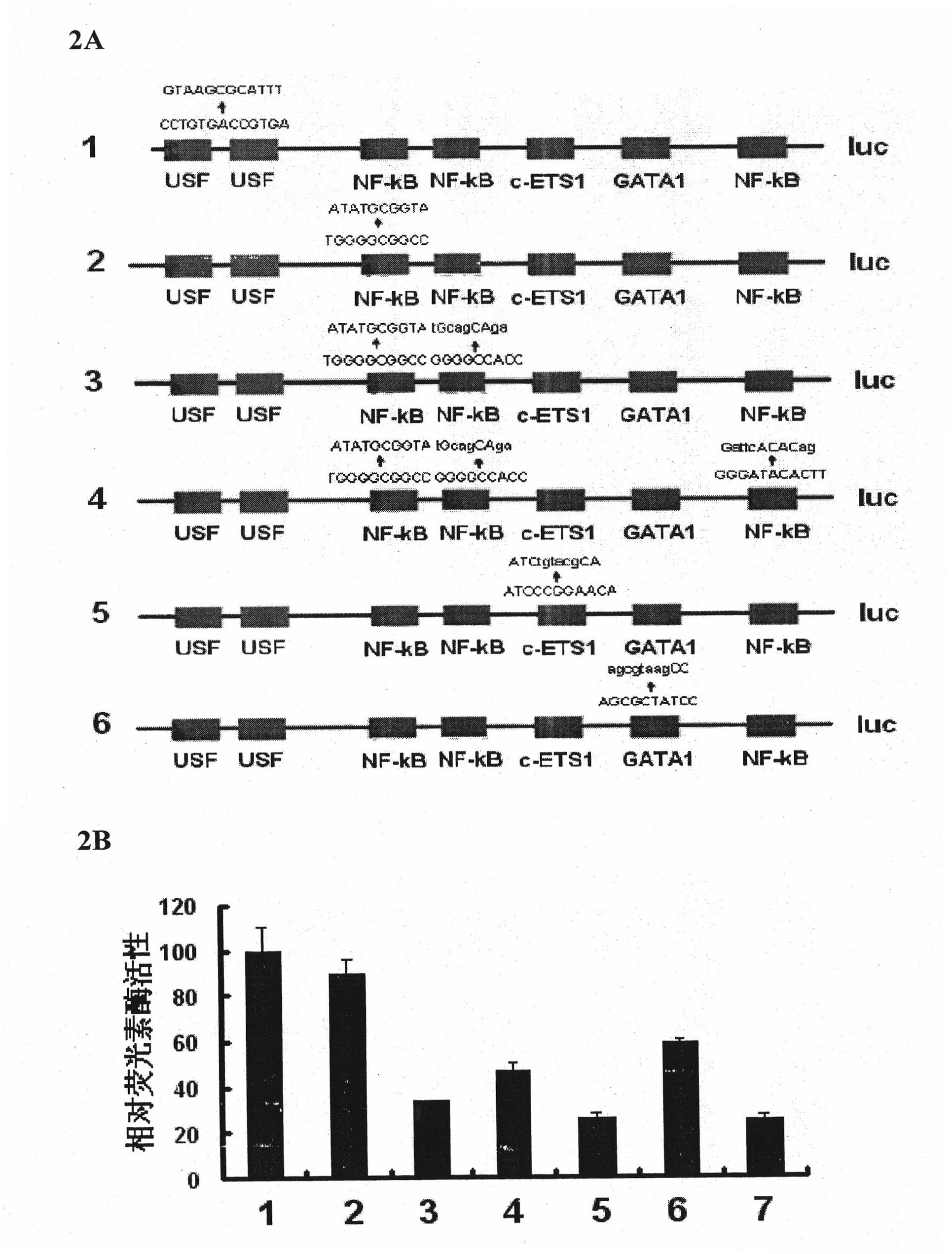
[0129] 2. Amplify the FAK promoter sequence and construct a reporter gene vector:
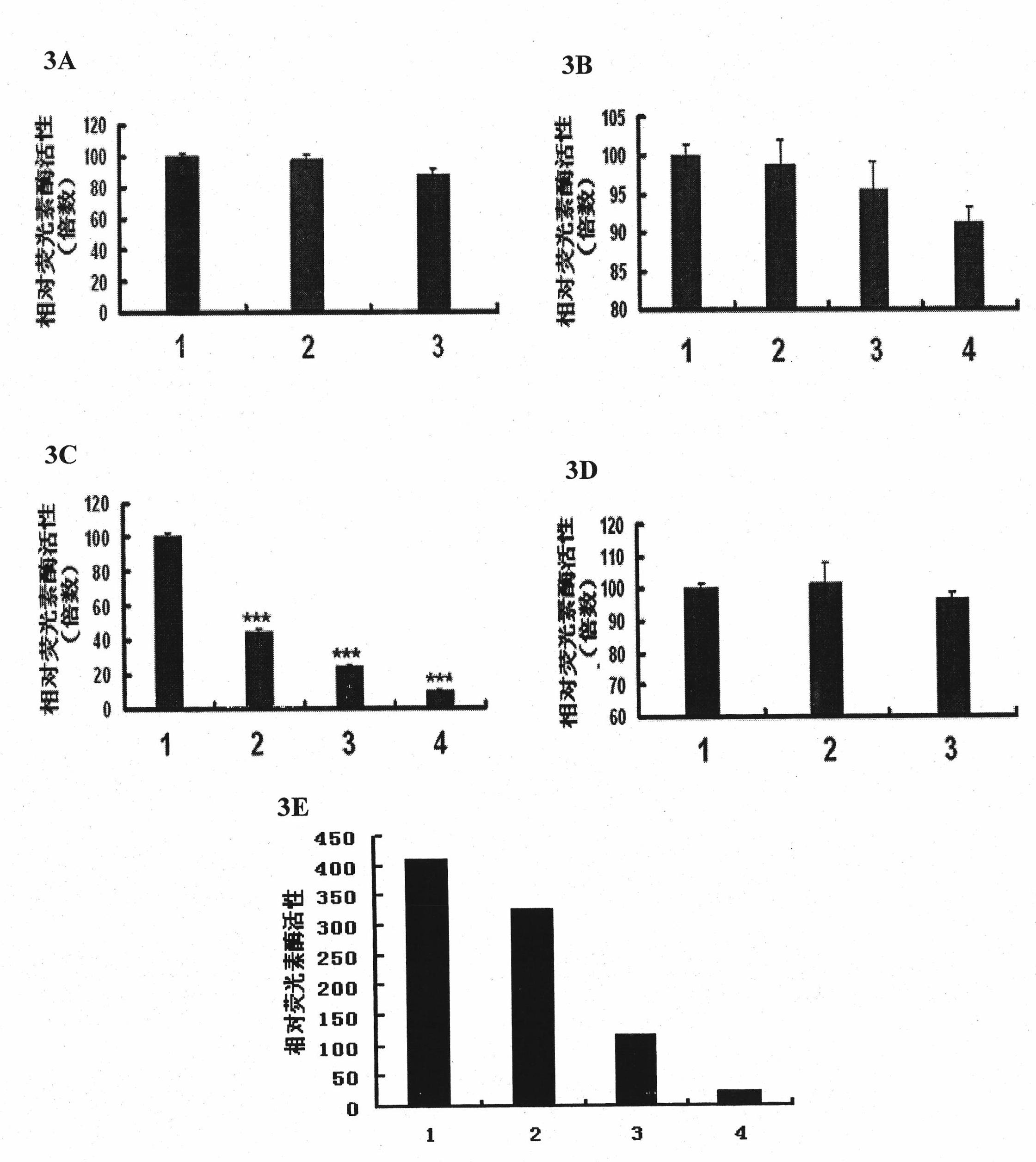
[0130] On the basis of the theoretical and experimental analysis of the mouse FAK promoter in Example 1, use the TRANSFAC database and bioinformatics methods to analyze and predict the human FAK gene promoter and its transcription factor binding site, and design and amplify the human FAK promoter Primers ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com