Method for detecting by using labeled probe and analyzing fusion curve
A technology of labeling and melting temperature, which is applied in the direction of biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problem of inability to perform melting curve analysis, inability to analyze the melting curve of mixtures, and difficulty in detecting the difference in probe fluorescence intensity, etc. question
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0083] Example 1: Design and synthesis of primers and probes
[0084] Rifampicin and isoniazid are the key first-line drugs in the tuberculosis treatment regimen and the anti-tuberculosis drugs most prone to drug resistance. Studies have shown that 97% of rifampicin-resistant strains are caused by mutations in rpoB genes (511, 513, 515, 516, 518, 519, 522, 526, 531 and 533 sites), and the mutations are mainly concentrated in a highly conserved 81bp region. Isoniazid resistance is mainly related to the katG gene (position 315) encoding catalase-peroxidase, the inhA gene encoding reduced nicotinamide adenine dinucleotide-dependent enoyl carrier protein reductase ( -8, -15) and mutations in the ahpC gene (-6, -9, -10 and -12) encoding alkyl catalase. The embB306 mutation is a highly specific genetic marker associated with tuberculosis multidrug resistance. Design primers and probes based on the above five gene sequences related to tuberculosis drug resistance, and ensure that ...
Embodiment 2
[0086] Embodiment 2: PCR reaction composition
[0087] Prepare the PCR reaction solution according to the following table (Table 1) and add the TB DNA template (each experiment must include a negative control and a wild-type positive control). After the preparation is completed, vortex and shake to mix well, and then put on the machine after centrifugation.
[0088] Table 1 Composition of each component of the PCR system (master MixI)
[0089] Component name
Embodiment 3
[0090] Embodiment 3: Reaction program setting
[0091] Set the fluorescence detection channel for collecting FAM fluorescence signals, put the PCR reaction tube into the fluorescent quantitative PCR instrument to start amplification, the reaction procedure is as follows (taking Rotor Gene Q as an example):
[0092] Table 10 PCR program settings
[0093]
[0094] Result judgment
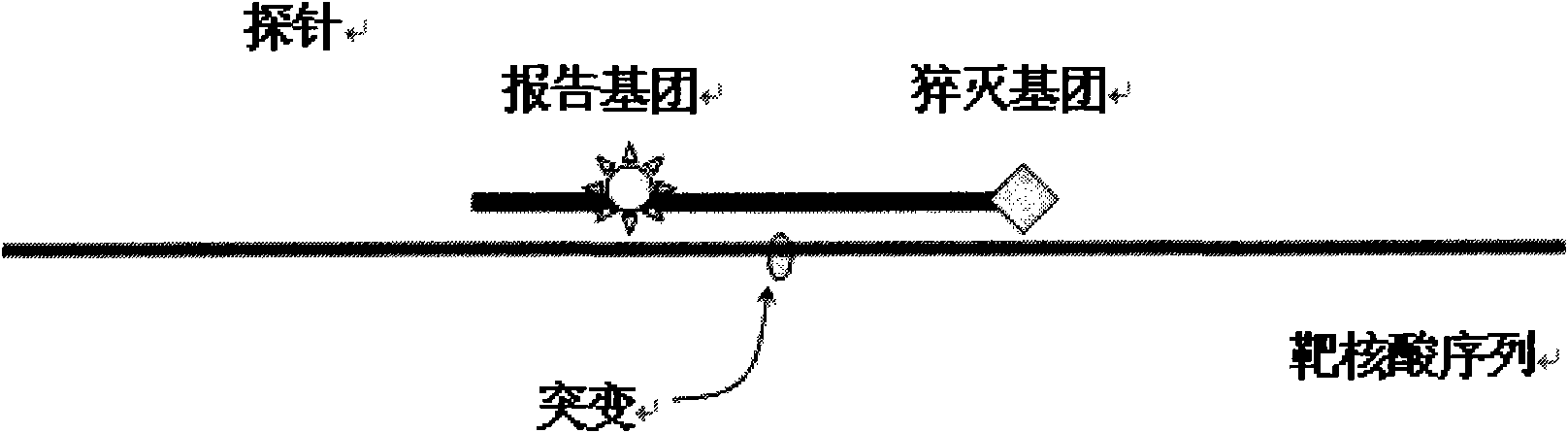
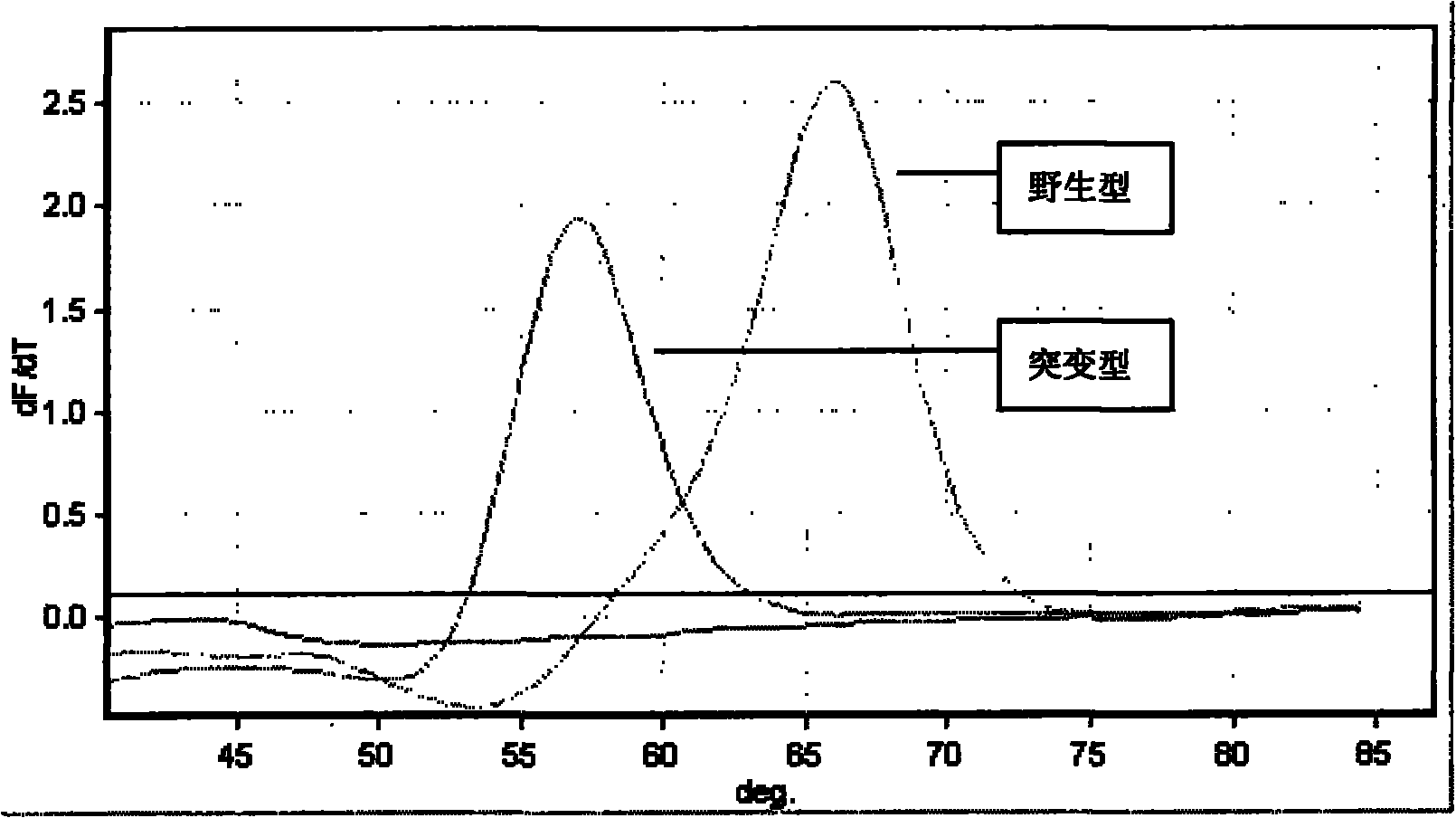
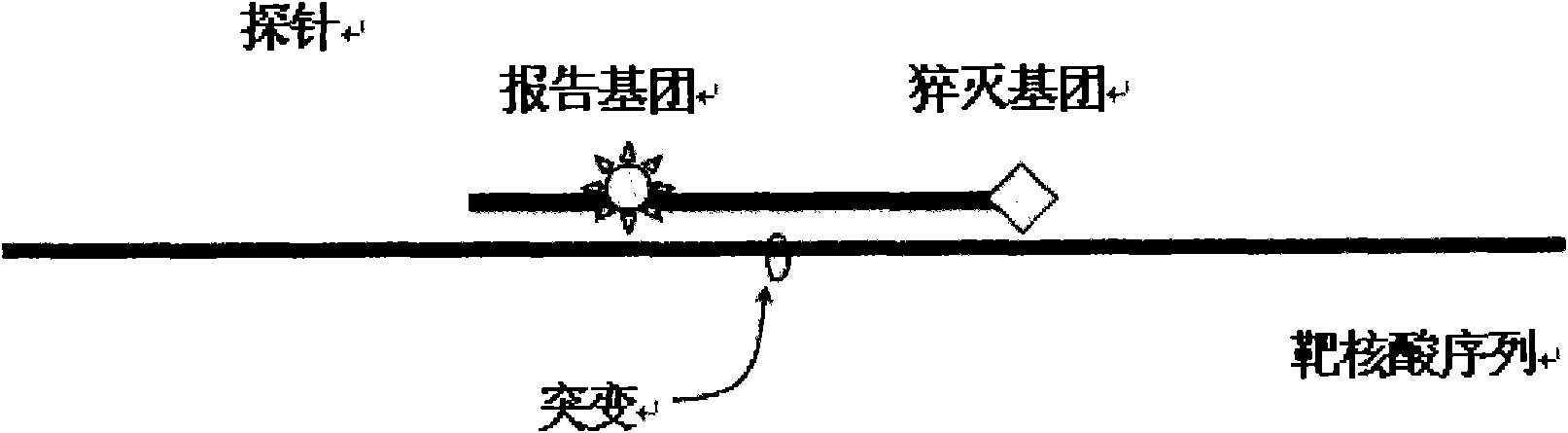
[0095]After the fluorescent quantitative PCR instrument runs, use its supporting software to analyze the melting curve of the results of this experiment. Taking the positive control product reaction tube (wild type) as a reference, the sample whose melting curve is exactly the same as the positive control product tube peak type is wild type; the sample whose melting curve is different from the positive control product tube peak type is the mutant type. When the gene is mutated, the Tm value of its melting curve will decrease compared with the wild type, the results are shown in the attached fig...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com