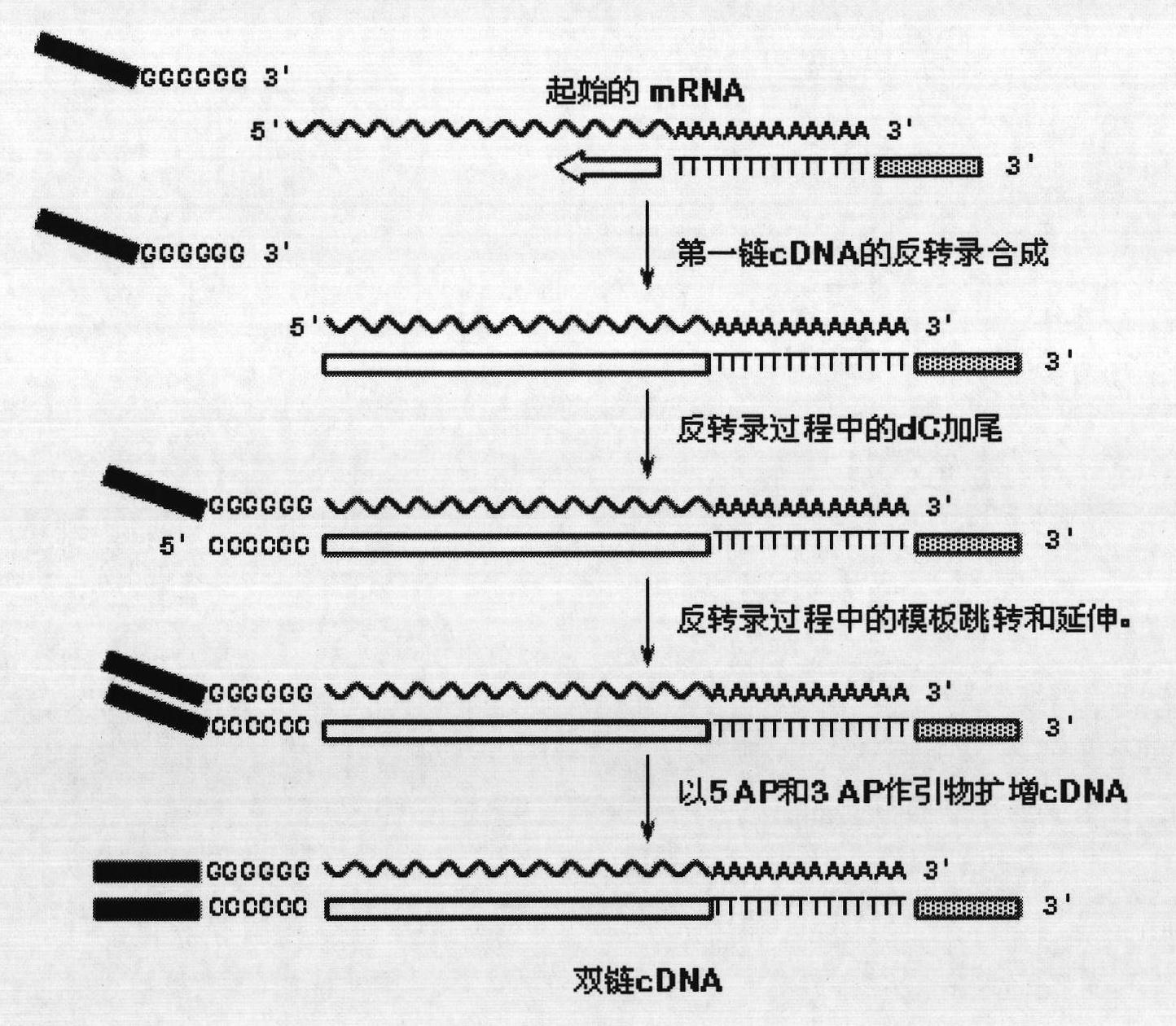
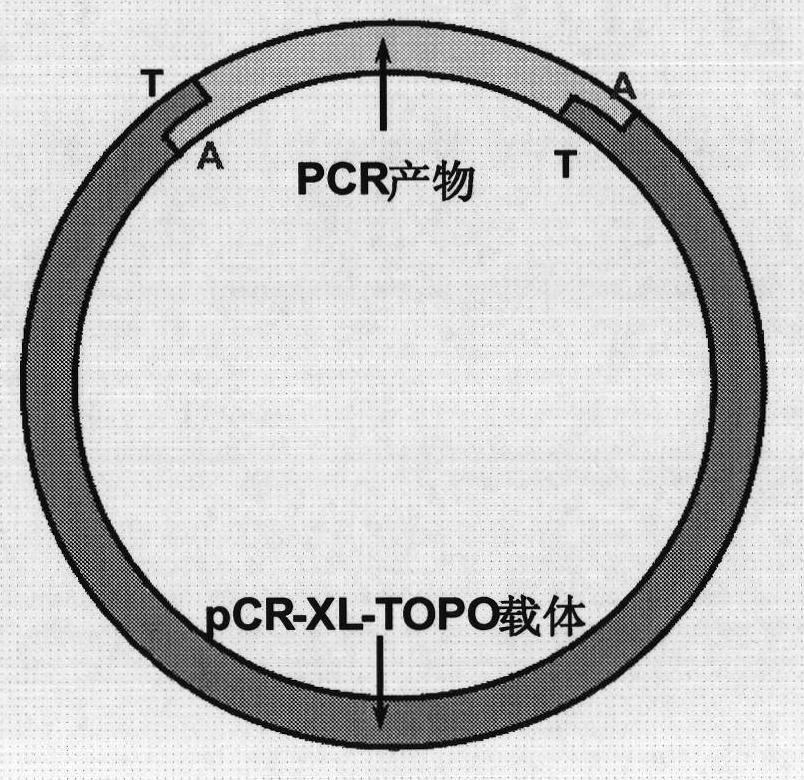
Method for constructing eukaryote cDNA library and special primers thereof
A eukaryotic library technology, applied in the field of eukaryotic cDNA library construction, can solve the problems of information loss, affecting the accuracy of results, etc., and achieve the effect of reducing difficulty
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Example 1: (Construction of cDNA library in peripheral blood of Holstein cows after heat stress treatment)
[0044] 2 mL of Holstein cow tail vein blood was collected with vacuum blood collection tubes (EDTA anticoagulant tubes) (BD). After heat-shock incubation at 42°C for 4 hours, total RNA was extracted from peripheral blood by Trizol (Invitrogen) method. 0.8 μg of total RNA was used as a template for reverse transcription, and SuperScript II reverse transcriptase (Invitrogen) was used for the reverse transcription reaction, and 3RRT and 5RRT primers were used to carry out the reverse transcription reaction of the sample total RNA. After the reverse transcription, 1 μL of the reverse transcription product was used as a template for PCR amplification, and the double-stranded cDNA was amplified with Ex Hot Start DNA polymerase (TaKaRa) and primers 3AP and 5AP. Use 1% agarose gel electrophoresis to separate the double-stranded cDNA, and cut the double-stranded cDNA with...
Embodiment 2
[0072] Embodiment 2: (tobacco leaf cDNA library construction)
[0073]Tobacco leaves were collected, and the total RNA of tobacco leaves was extracted by Trizol (Invitrogen) method. 4.8 μg of total RNA was used as a template for reverse transcription, and SuperScript II reverse transcriptase (Invitrogen) was used for the reverse transcription reaction, and 3RRT and 5RRT primers were used to carry out the reverse transcription reaction of the sample total RNA. After the reverse transcription, 1 μL of the reverse transcription product was used as a template for PCR amplification, and the double-stranded cDNA was amplified with Ex Hot Start DNA polymerase (TaKaRa) and primers 3AP and 5AP. Use 1% agarose gel electrophoresis for electrophoretic separation of double-stranded cDNA, and cut double-stranded cDNA with a molecular weight of 500-5000bp in the agarose gel to remove small fragment products less than 500bp or incomplete synthesis; use Omega's E.Z.N. The gel recovery kit i...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com