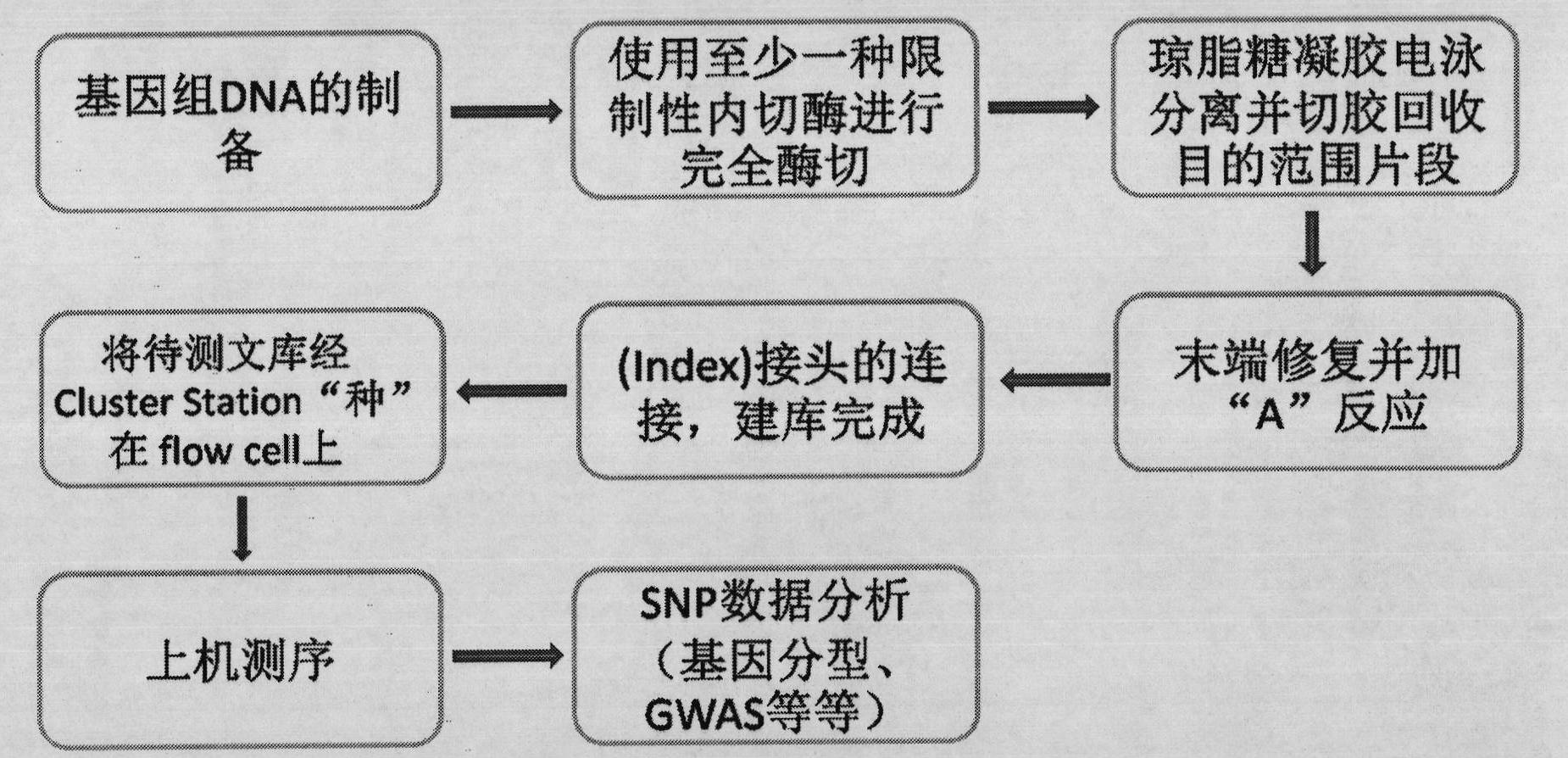
DNA (deoxyribonucleic acid) library and preparation method thereof as well as method and device for detecting single nucleotide polymorphisms (SNPs)
A DNA library and detection method technology, applied in the field of molecular biology, can solve cumbersome and other problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
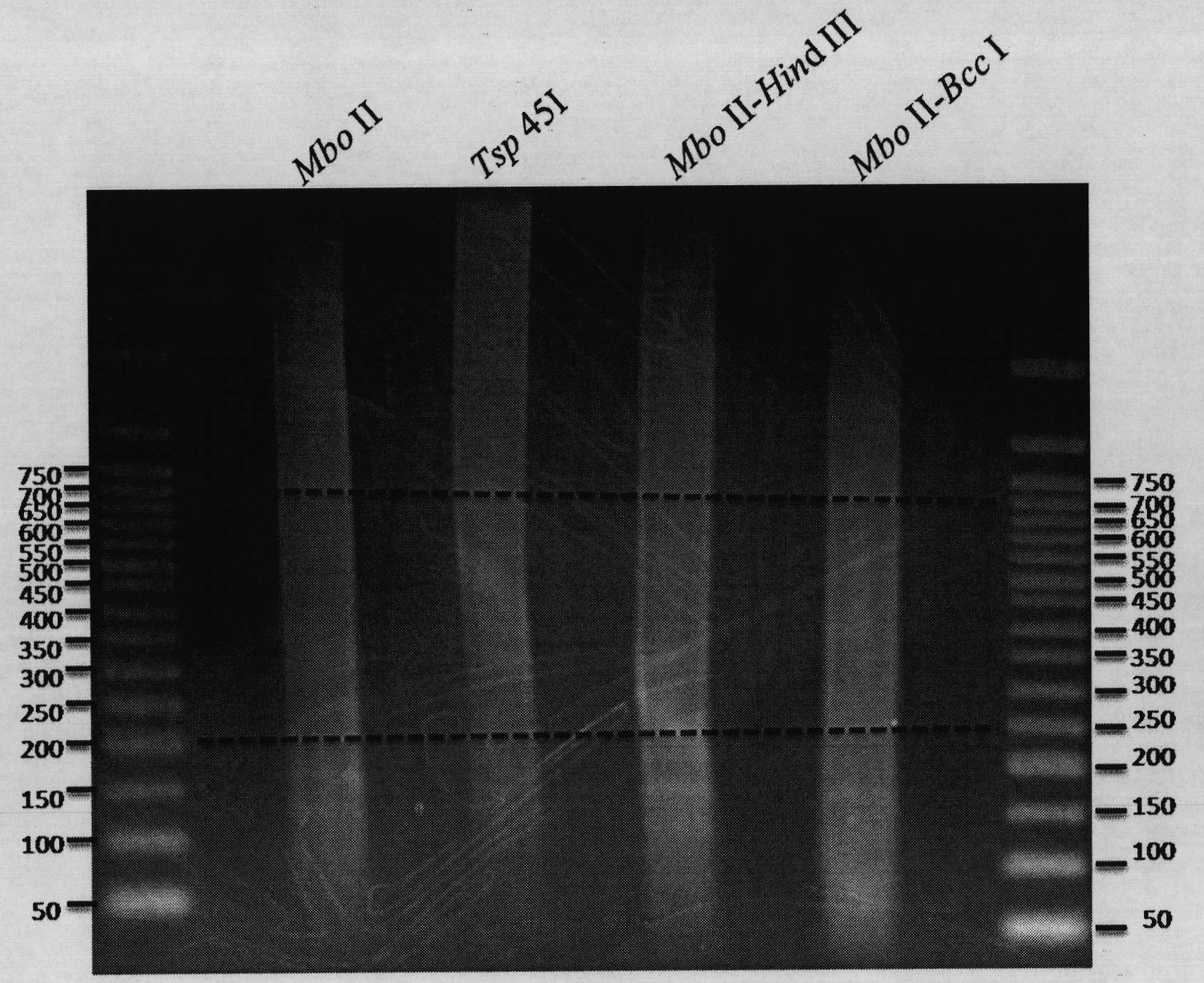
[0072] Example 1: Determination of preferred restriction enzymes or enzyme combinations
[0073] According to the recognition sequence of the enzyme or combination of enzymes in Table 1, through the known restriction recognition site information, the hg18 genome sequence is used as the reference sequence, and the restriction site is used as the boundary to classify the genome according to the length range, and finally select 200bp- Fragments in the range of 700bp were used as the library collection to be tested. For those skilled in the art, the hg18 genome sequence data can be downloaded from known databases, for example, from http: / / genome.ucsc.edu / .
[0074] according to HiSeq2000 TM The PE91 index sequencing parameters were used to filter the generated data. Since PE91 cycle number sequencing is used in actual sequencing, the 91bp bases at both ends of each fragment in the above library collection are used as the target region, so that the fragments within the select...
Embodiment 2
[0083] Example 2: Sequencing of Yanhuang No. 1 DNA Library
[0084] For the human genome, such as the preferred enzyme digestion combinations expressed in the detailed technical method Table 1, select four optimal enzyme digestion combinations in which the recovered fragments are in the range of 200bp-700bp for enzyme digestion library construction, through data analysis and Table 1 compared to the results shown. The specific operation is as follows:
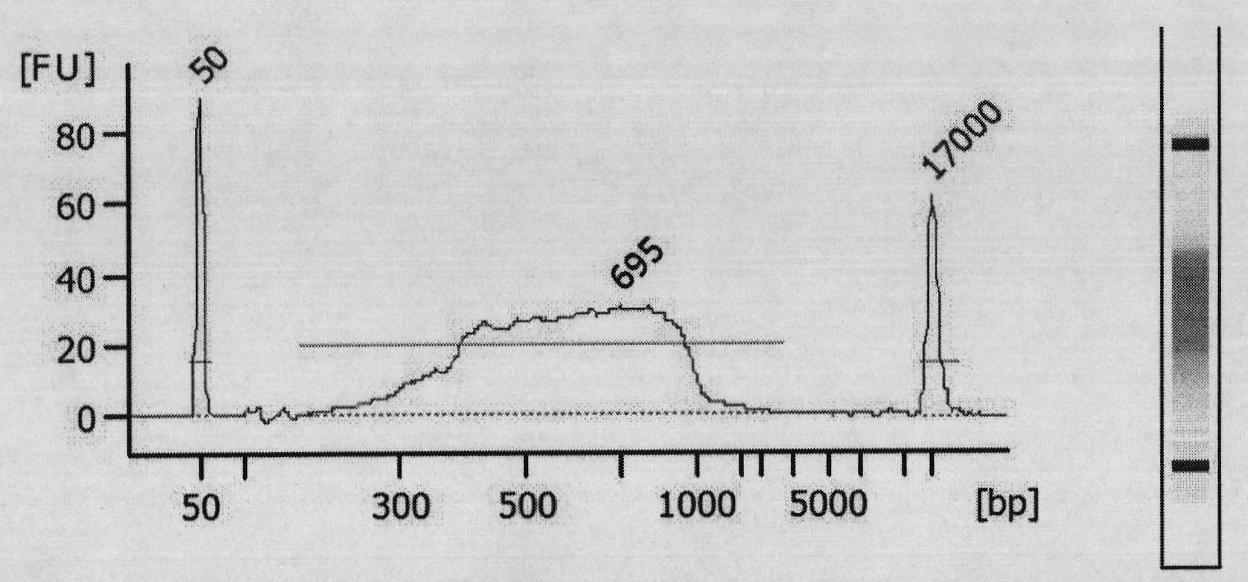
[0085] Human genomic DNA is extracted from the blood cells of Yanhuang No. 1 (YH1), which is extracted using The DNA Mini Kit (QIAGEN) was completed, and the operation was carried out completely according to the instructions. Finally, the genomic DNA was dissolved in EB buffer, and After ND-1000 was quantified by the absorbance value at A260, 5 μg was taken for enzyme digestion. All restriction endonucleases were purchased from NEB Company, and the buffer solution was provided with the enzymes. A total of four combinatio...
Embodiment 3
[0117] Example 3: SNPs detection and genotyping using Tsp 45I digestion to build a library
[0118] In order to detect the parallelism between different samples and the actual detection of SNPs, in addition to the Yanhuang No. 1 (marked as YH) genome, another healthy male (marked as DY) genome was selected for parallelism in this example. experiment. According to the method similar to that in Example 2, two DNA libraries were constructed with Tsp 45I enzyme: YH library (YH library trial2) and DY library.
[0119] The detection of SNPs uses the SOAPsnp program to filter according to the filter parameters of Q20. mean quality of bestallele > 20. copy number ≤ 1.1. Finally, the actual number of SNPs obtained and the proportion of these sites in the dbSNP database are counted. At the same time, according to the existing SNP site information of the whole genome of Yanhuang No. 1 (Ruiqiang Li et al., (2010). SNP detection for massively parallel whole-genome resequencing. Genome R...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com