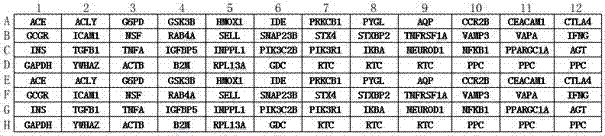
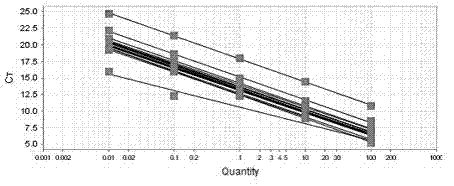
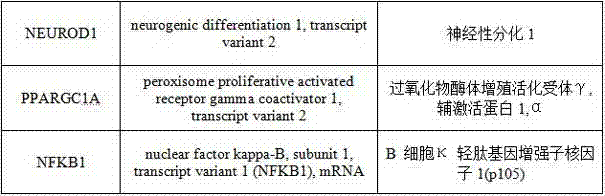
T2DM (Type 2 Diabetes Mellitus) detection primer group, PCR (Polymerase Chain Reaction) chip and detection method for human and monkeys
A type 2 diabetes detection chip technology, applied in the field of fluorescent quantitative PCR detection chip, can solve the problems of failure to obtain primer sets and other problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0057] Example 1. Extraction of white blood cells.
[0058] ① Collect venous blood in the anticoagulant tube and mark each other; the samples 1-5, 26-32 health monkey samples; the sample 6-10 is the crab monkey sample of the initial stage of the model; the sample 11-15 is the model of the crab monkeys in the model;Sample 16-20 clinical early clinical crab and monkey sample; sample 21-25, 33-39 is a clinical crab monkey sample; sample 40-46 is a healthy sample; sample 47-53 is a diabetic sample.
[0059] ② Add 1ml lymphocyte extract to the 25ml centrifugal tube.
[0060] ③ Slowly add an equal volume of PBS buffer (1 ×) to the anticoagulant tube equipped with 10ml blood, mix well.
[0061] ④ Slowly add the mixture of ③ ② to ② with a pipetter, and try not to sink the mixed liquid into the bottom of the ②, causing it to suspend it on the lymphocyte separation liquid.
[0062] ⑤ Corporation on frozen centrifuge, 4 ℃, 1700g, 15min.
[0063] ⑥ Take out the centrifugal tube and carefully ...
Embodiment 2
[0066] Example 2. Extract of RNA.
[0067] ① Take out the leukocytes obtained from the implementation example 1 from the liquid nitrogen. The room temperature is static. The finger wall of the bomb pipe can be scattered. The 1ml Trizol is added to each frozen tube to fully mix.
[0068] ② Layers: After the sample is added to Trizol, the room temperature is placed for 5 minutes to make the sample split.4 ° C, 15000g centrifugal 10min take the Qing.Add 200 μL of chloroform in Shangqing. After the severe oscillation is mixed, the room temperature is placed for 3-5min.
[0069] ③ RNA precipitation: 4 ℃, 15000g centrifugal 10-15min.The sample will be divided into three layers: yellow organic phase, the middle layer and colorless water phase, and the RNA is mainly in the water phase, and the water phase (usually 550 μL can be absorbed) to the new tube.Be careful to absorb the water phase. Do not absorb the intermediate interface, otherwise it will cause DNA pollution in RNA samples.Add ...
Embodiment 3
[0072] Example 3.CDNA reverse transcription.
[0073] Use Transgene's CDNA transliteration kit-Easyscript First-Strand CDNA SYNTHESIS SUPERMIX (TRANSGEN BIOTECH, AE301) to transcrIn the system, the total RNA volume is about 0.25-0.5 μg.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Upstream primer | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com