Microfluidic-based nucleic acid hybridization reaction platform and hybridization analysis method
A technology of nucleic acid hybridization and microfluidics, which is applied in specific-purpose bioreactors/fermenters, biochemical equipment and methods, bioreactor/fermenter combinations, etc., can solve the problem of large actual consumption and achieve sample consumption Few, easy integration, and short hybridization times
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
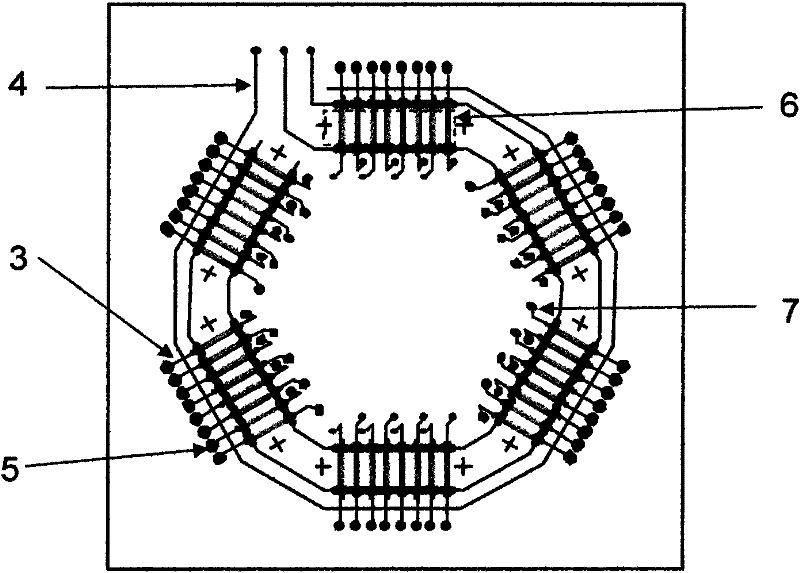
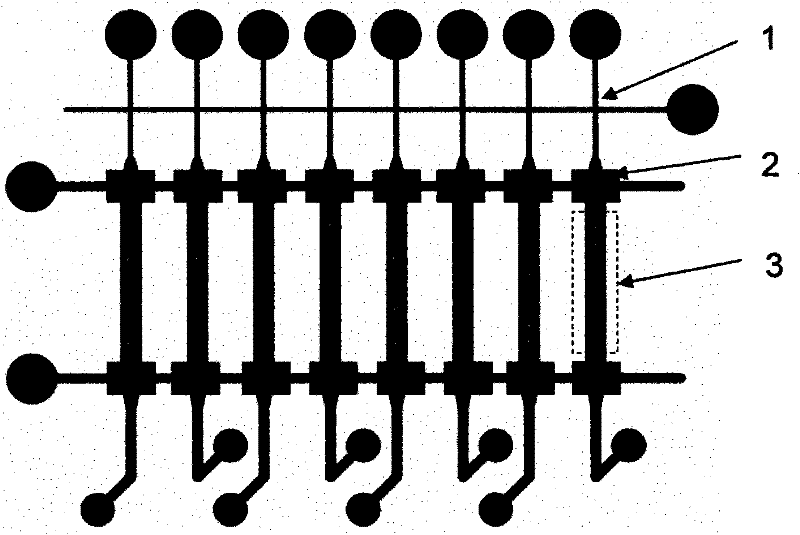
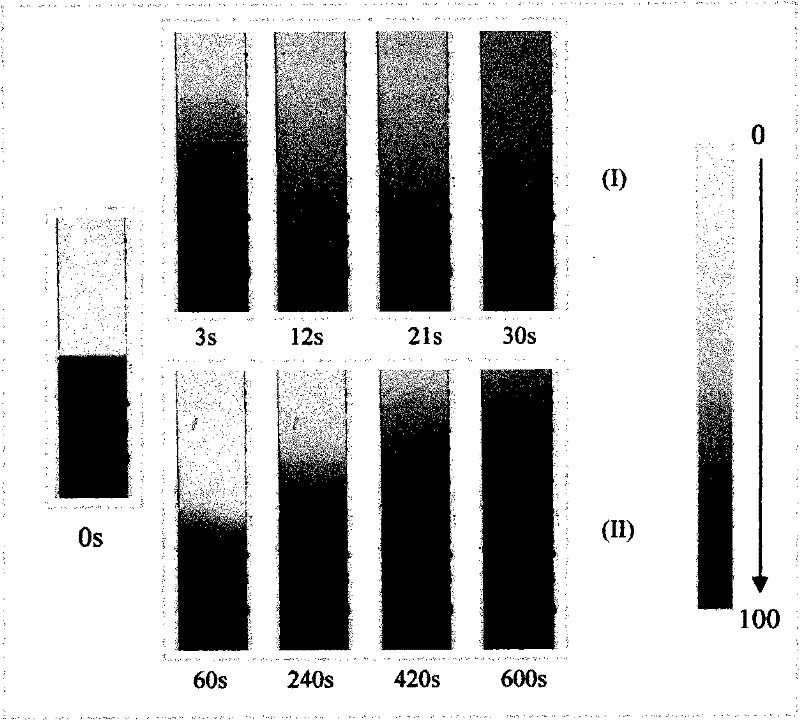
[0027] The overall diagram of the chip device of the microfluidic nucleic acid hybridization platform based on micropump and microvalve control is shown in figure 1 , the schematic diagram of its 8-channel hybridization analysis functional unit is shown in figure 2 . First, 1 uL of DNA sample was added to the sample chamber in advance. The sample enters the hybridization channel through capillary force or negative pressure, and the program controls the upper air circuit to repeatedly apply positive pressure and negative pressure at both ends of the hybridization area, and the liquid flow shuttles back and forth in the channel under the pressure, continuously The time is 90-300s. After the hybridization reaction is finished, the waste liquid in the sample chamber and the channel is pumped out to the waste liquid chamber under the action of negative pressure. Then add washing liquid into the sample chamber, the capillary force or negative pressure makes the washing liquid en...
Embodiment 2
[0029] On the basis of the above functional units, the simultaneous rapid detection of 48 samples was carried out. Four different probe molecules are immobilized in each microfluidic channel, and the whole chip consists of 48 microfluidic channels. First, under the action of positive pressure, the microvalve is closed, and different DNA samples are added to each sample chamber; then the microvalve is opened, and the sample enters the microfluidic hybridization channel through capillary force or negative pressure, and the program controls the hybridization channel. The micropump at the end of the micropump continuously and repeatedly applies positive pressure and negative pressure, and the liquid flow shuttles back and forth in the channel under the action of pressure for 90s; The hybridization spots were washed for 30 s, and the signal was detected under a fluorescence microscope. Such as Figure 7 As shown, the simultaneous detection of 48 samples is realized on the inventi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com