Method for detecting exon 19 deletion mutation and exon 21 point mutation of epidermal growth factor receptor gene
A technology of epidermal growth factor and deletion mutation, applied in the field of molecular biology, can solve the problems of shortened time for results, high price, and limitations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0066] Embodiment 1 Primer and probe design
[0067] 1. Materials
[0068] Restriction endonucleases Msel and Mscl were purchased from NEB Company in the UK, and 2×Universe Taqman Mix was purchased from ABI Company in the United States (this Mix already included PCR buffer, dNTP, Mg 2+ , all provided by the supplier), the primers and probes used were synthesized by Shanghai Sangong, and the Tiangen tissue sample genomic DNA extraction kit was purchased from Beijing Tianhe Biotechnology Co., Ltd. The instrument used was ABI7300 fluorescent quantitative PCR instrument from ABI Company of the United States.
[0069] 2. Primer and probe sequence design
[0070] 1. Enrichment primer design
[0071] Primers that can amplify a section of 400-500bp were designed respectively, and the relevant parameters were: Tm value 65.0°C-68.0°C, GC value 40.0%-65.0%, primer size 23±3bp. The designed enrichment primer sequences are as follows:
[0072] E746_A750del upstream primer: 5'-CTTCCAAA...
Embodiment 2
[0086] Example 2 Using this method to detect E746_A750del and L858R mutations
[0087] The mutation-enriched ARMS fluorescent quantitative PCR detection method of the present invention integrates enzyme digestion enrichment PCR technology and ARMS fluorescent quantitative PCR technology, that is, samples are subjected to enzyme digestion, PCR enrichment and ARMS fluorescent quantitative PCR identification in one reaction system. . The specific process is:
[0088] 1. Sample preparation: Genomic DNA was extracted using Tiangen Tissue Sample Genomic DNA Extraction Kit.
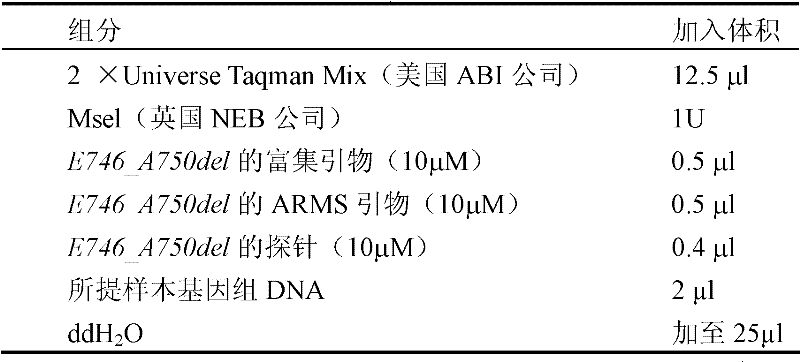
[0089] 2. Detection method: first, the samples and the negative control were incubated in 25 μl reaction system at 37°C for 30 minutes, and the endonuclease Msel (NEB, England) or Mscl (NEB, England) was used to treat EGFR wild-type exon 19 respectively. Or 21 for enzyme digestion to reduce the interference of wild-type DNA on mutated bases. Then carry out the PCR reaction and run the first cycle to enrich the ...
Embodiment 3
[0097] Example 3 Clinical tissue sample detection
[0098] 1. Materials and methods
[0099] A total of 73 lung cancer tumor tissue samples were collected from Union Medical College Hospital and China Institute for the Identification of Pharmaceutical and Biological Products, and the whole genome DNA was extracted using the Tiangen Tissue Sample Genomic DNA Extraction Kit.
[0100] 2 μl each was taken for mutation enrichment ARMS fluorescent quantitative PCR detection, and sterile deionized water was used as a negative control. See Example 2 for specific implementation methods.
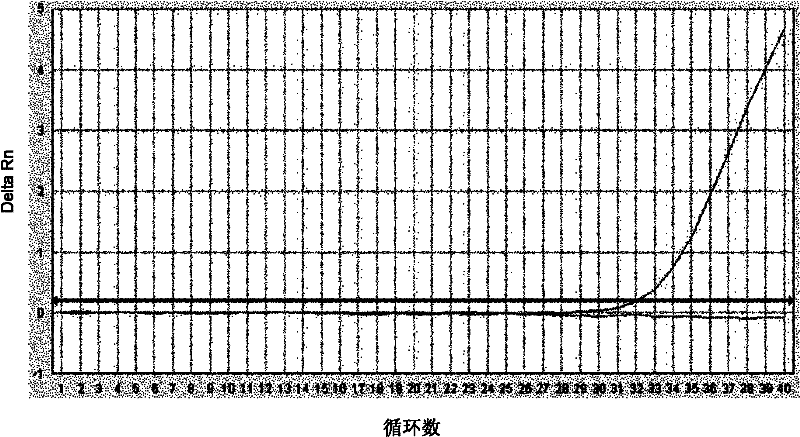
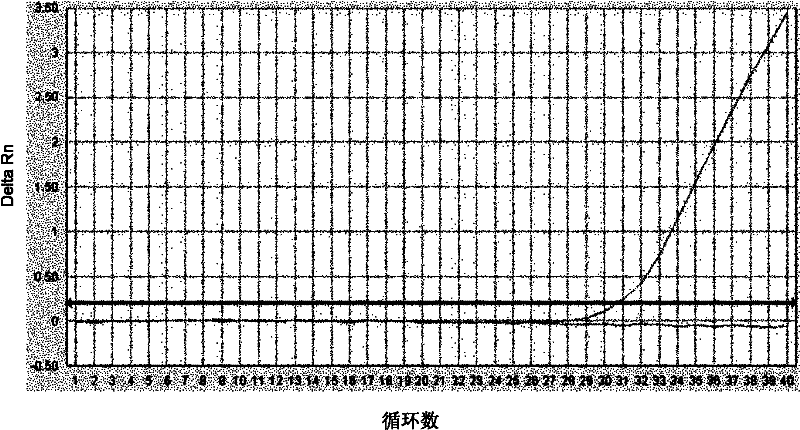
[0101] 2. Experimental results
[0102] In 73 tissue samples, 12 E746_A750del deletion mutations and 11 L858R mutations were detected. The detection method is determined to be the same as in Example 2.
[0103]
[0104]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com