Primers and probe for detecting mouse Sendai virus and method thereof
A Sendai virus and probe technology, applied in the field of biological detection, can solve the problems of short detection time and achieve the effects of simple and fast operation, improved sensitivity, and good linear relationship
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0039] The design of embodiment 1 mouse Sendai virus special primer and probe
[0040] Sendai virus RNA is composed of 15383 nucleotides, encoding 6 viral structural proteins and one non-structural protein. The 6 structural proteins are L (large), P (RNA polymerase), NP (nucleocapsid), HN (hemagglutinin-neuraminidase ), F (Fusion) and M (membrane). A nonstructural protein, C, is unique to viruses. The gene sequence is 3'-Leader-NP-P+C-M-F-HN-L-Leader-5', with a leader sequence at the 3' end and 5' end.
[0041] By comparing the genomes of different strains of SEV in GenBank, the conserved homologous sequence in the non-structural protein of the virus is selected: the nucleotide sequence shown in SEQ ID NO:4.
[0042] For the found conserved homologous sequence region, special primers and probe sequences for real-time fluorescent quantitative PCR detection were designed, and the amplified target fragment was 100 bases in size. Wherein the preferred primer and probe sequences...
Embodiment 2
[0049] The preparation of embodiment 2 positive RNA standard items
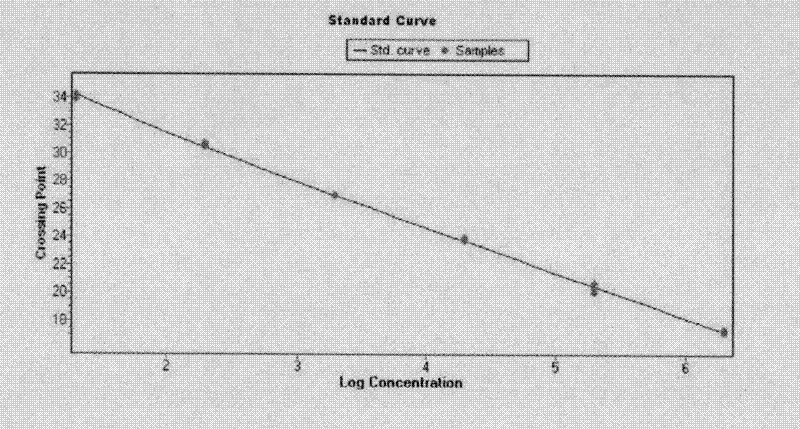
[0050] First prepare the pMD18-T vector containing the nucleotide sequence shown in the homologous sequence SEQ ID NO: 4, use it as the vector to amplify a 100bp DNA fragment, recover it for in vitro transcription reaction, digest the DNA template and purify the RNA after the reaction , quantified as a positive RNA standard.
[0051] 1. Preparation of templates for in vitro transcription
[0052] The nucleotide sequence shown in SEQ ID NO: 4 was synthesized and inserted into the pMD18-T vector (synthesized by Invitrogen), and named pMD18T-SEV. Using pMD18T-SEV as a template, use the upstream primer SEV-100bp-F: 5'-TAATACGACTCACTATAGGGCATCTATG-3' and the downstream primer SEV-100bp-R: 5'-TTTGCAAACACAATAC-3' to amplify a 100bp DNA fragment , recovered by agarose gel electrophoresis, used as a template for in vitro transcription.
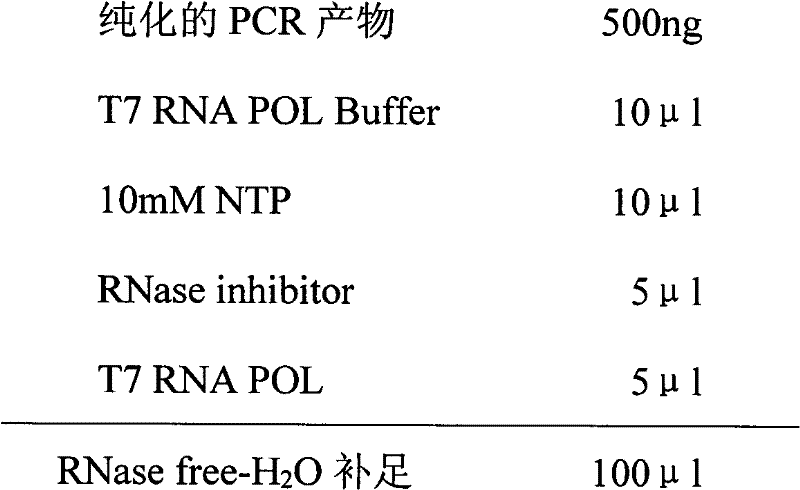
[0053] 2. In vitro transcription to prepare a large amount of RNA
[0054] Pr...
Embodiment 3
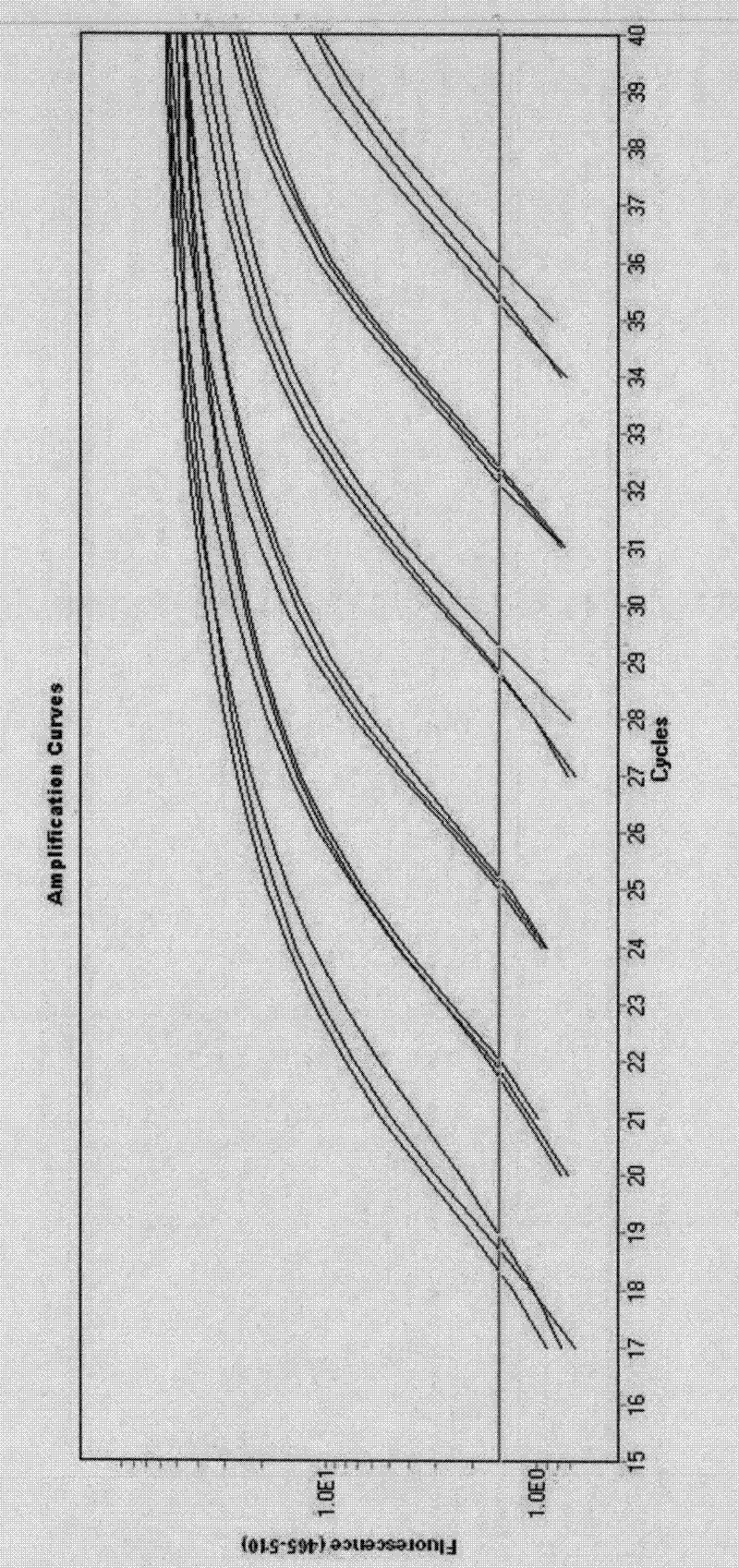
[0066] Embodiment 3 Establishment of real-time fluorescent quantitative PCR amplification method
[0067] 1. Real-time fluorescence quantitative PCR template preparation - total RNA extraction:
[0068] According to the instructions for use, the sample RNA was extracted with Trizol / Trizol LS kit (provided by Invitrogen), the specific method is as follows:
[0069] 1) Add an appropriate amount of TRIzol / / Trizol LS to the sample (if it is a tissue that needs to be ground and homogenized), mix well, and let stand at room temperature for 5 minutes;
[0070] 2) Add 200 μl of chloroform: isoamyl alcohol (24:1) solution, mix vigorously with vortex for 10 seconds, leave at room temperature for 5 minutes, and centrifuge at 15,000 rpm at 4°C for 15 minutes;
[0071] 3) Take the supernatant, add an equal volume of isopropanol to precipitate, and centrifuge at 15,000 rpm at 4°C for 15 min;
[0072] 4) Remove the supernatant, add pre-cooled 75% ethanol (volume ratio 1:1) to wash the RNA ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com