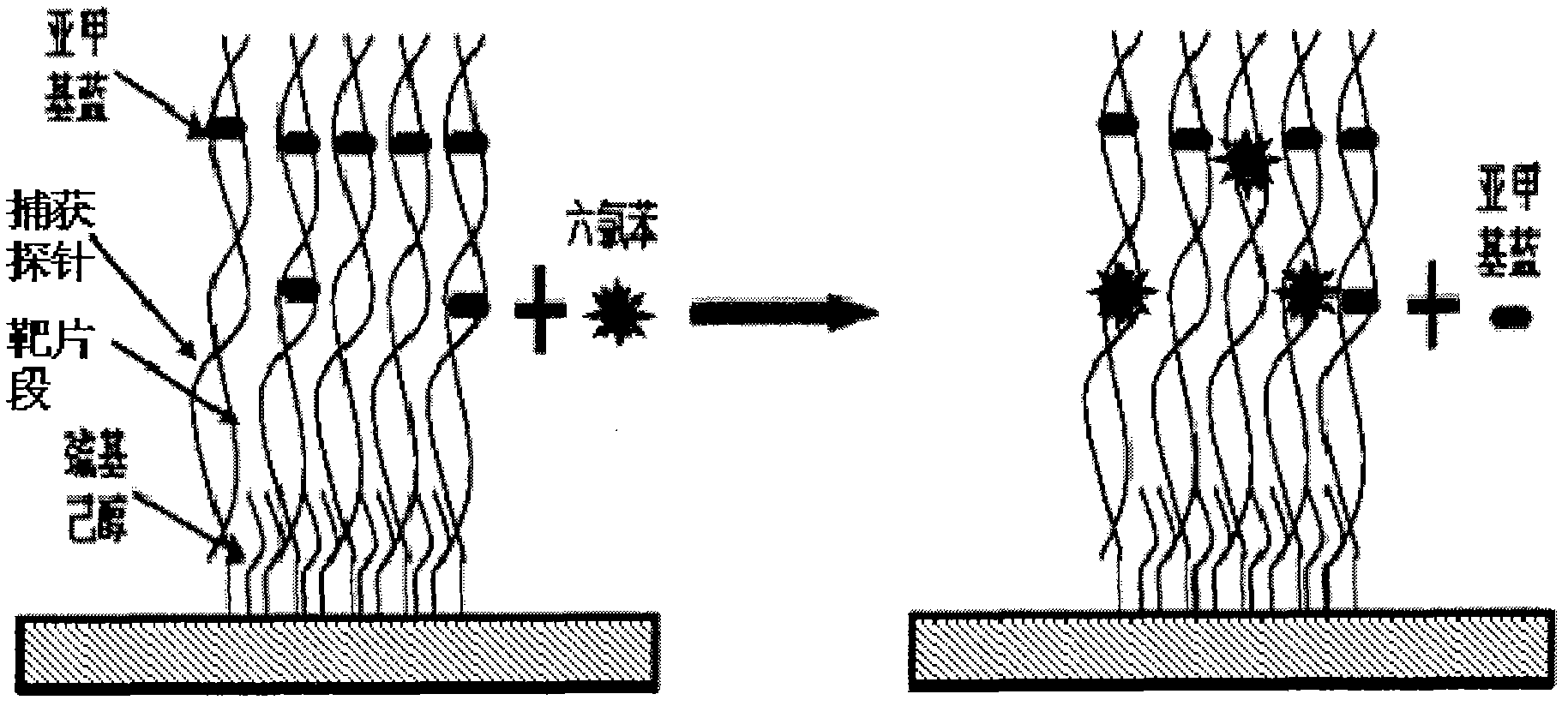
Electrochemical method employing DNA as probe to detect environmental pollutant
A detection environment, electrochemical technology, applied in the field of biochemical analysis and biosensing, can solve the problems of insignificant changes in electrical signals, reduce instrument sensitivity, low response signal, etc., to achieve expanded detection range, reliable detection results, and simple equipment Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Example 1. Nucleic acid biosensor assembly
[0043] Gold electrode assembly steps:
[0044] 1) The surface area is 0.02cm 2 The gold electrodes were polished with Al2O3 powders with particle sizes of 1, 0.3, and 0.05 μm, and then ultrasonically cleaned three times in absolute ethanol and deionized water for 2 minutes each time. Put it into 1M sulfuric acid solution and scan and activate it cyclically between 0 and 1.7V. When the scanning curves are completely overlapped, stop scanning (activation is complete).
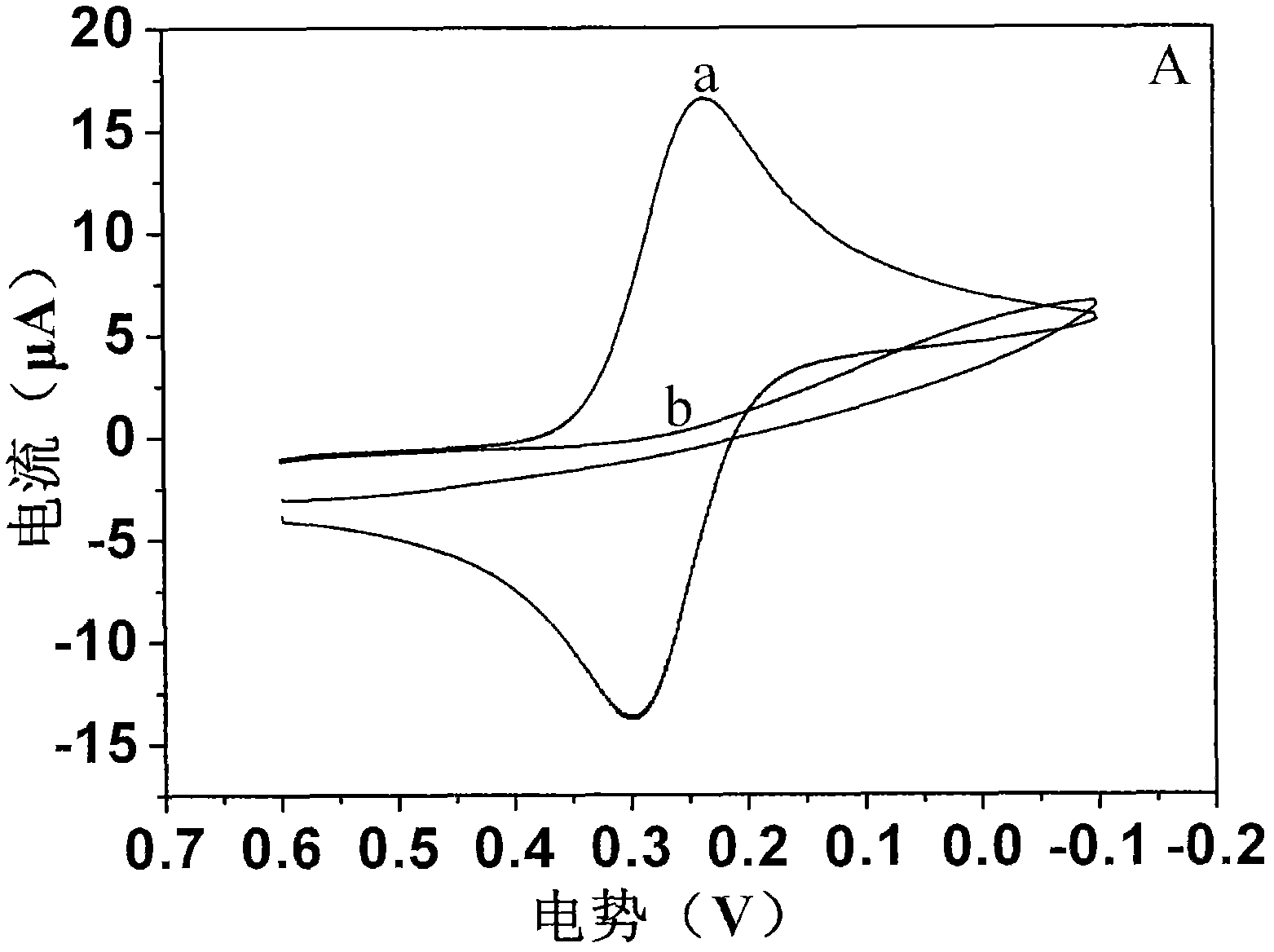
[0045] 2) figure 2 The cyclic voltammetry curve a in A indicates that the activated gold electrode is placed in 2mM potassium ferricyanide buffer solution, and the cyclic voltammetry curve is scanned between -0.1V and 0.6V. The curve redox peak potential difference is less than 75mV, indicating that the potassium ferricyanide reaction on the surface of the gold electrode is a completely reversible reaction, and the surface of the gold electrode has no impuri...
Embodiment 2
[0048] Embodiment 2. Nucleic acid biosensor detects hexachlorobenzene
[0049] Hexachlorobenzene was dissolved in 10 mM PBS buffer solution with N, N-dimethylformamide as co-solvent to prepare 100 pM, 1 nM, 10 nM, 100 nM and 1 μM hexachlorobenzene solutions.
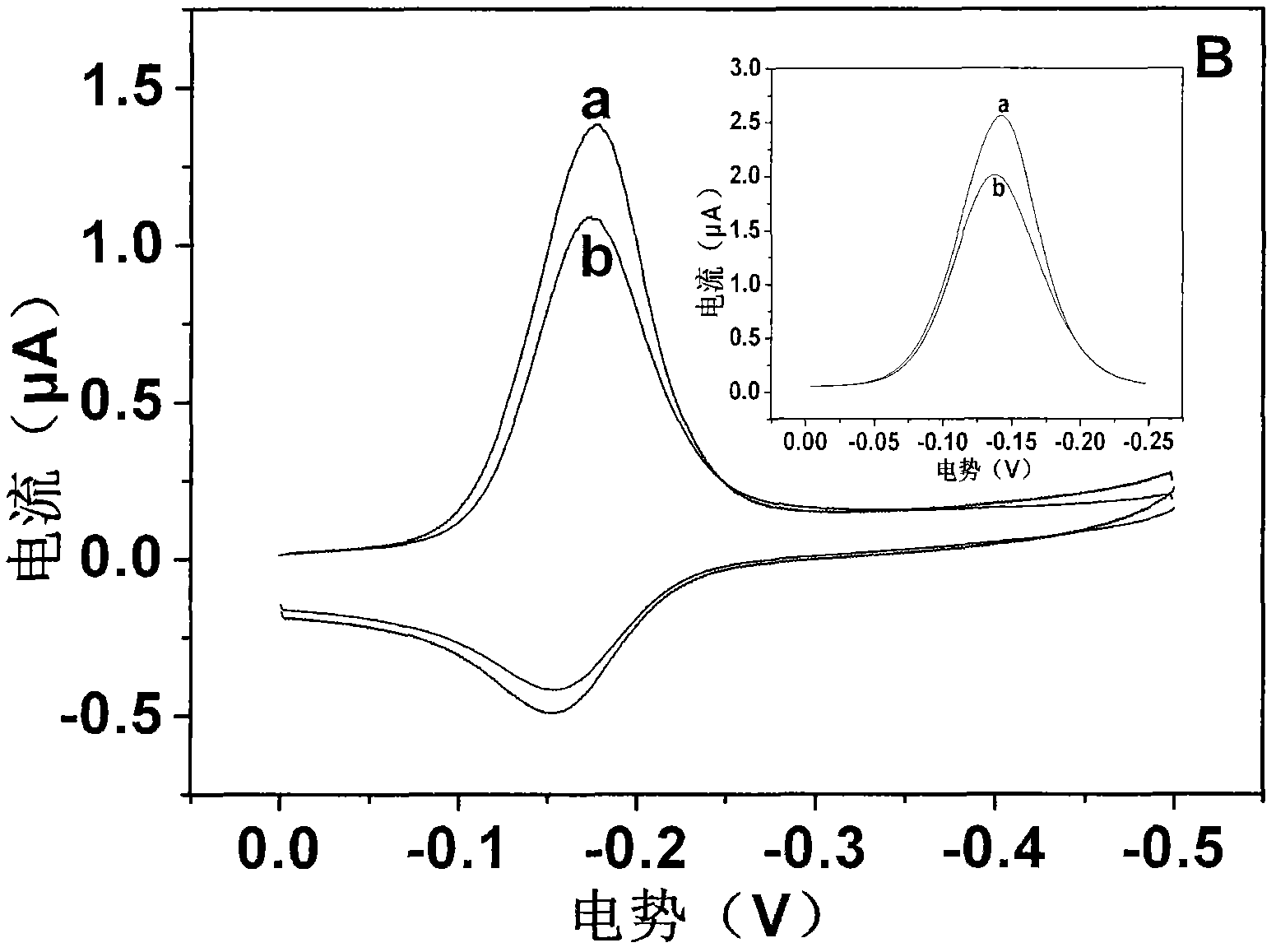
[0050] Figure 4 Representation: Add 15 μl of hexachlorobenzene solution dropwise to the surface of double-stranded DNA-modified gold electrode, treat it at room temperature for 20 minutes, then immerse in 10 μM MB of PBS (10 mM) buffer solution and scan electrochemically under nitrogen atmosphere to obtain the relationship between the concentration and the electrical signal related curves. Hexachlorobenzene interacts with DNA, causing changes in DNA pairing through non-covalent binding to double-stranded DNA, DNA base damage, etc., resulting in changes in the electrical signal of the nucleic acid sensor. As the concentration increases, the electrical signal gradually decays, and the intensity of the signal change is l...
Embodiment 3
[0051] Example 3. Ultraviolet-visible detection of the mode of action of hexachlorobenzene and DNA
[0052] Figure 5 Indicates: add 2ml of 10mM PBS buffer solution to the quartz cuvette, scan the blank at full wavelength within the range of 190-700nm, curve a is adding 20μl of 1mM MB to the blank solution, and scan at full wavelength to get the MB peak. Curve b is the mixed peak of MB and DNA obtained by adding 6 μl of 100 μM double-stranded DNA to the MB solution, shaking well and standing for 10 minutes. Select the non-interference peak at 620-700nm as the detection wavelength range. Curve c is adding 2μl 10 to the mixed solution of MB and DNA -2 M hexachlorobenzene, measure the absorption peak of the solution at 620-700nm.
[0053] According to the analysis of the experimental results, the MB peak appears at 190-350nm and 620-700nm, while the DNA peak is at 220-270nm, which interferes with the MB 190-350nm peak, so it is not suitable for analysis and research. The peak...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com