Rice Rhizoctonia solani effector gene AG1IA10434 and application thereof
A technology of AG1IA10434 and Rhizoctonia solani, applied in the field of genetic engineering, can solve problems such as unclear action sites of Rlm resistance genes, and achieve the effect of controlling the occurrence of sheath blight
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
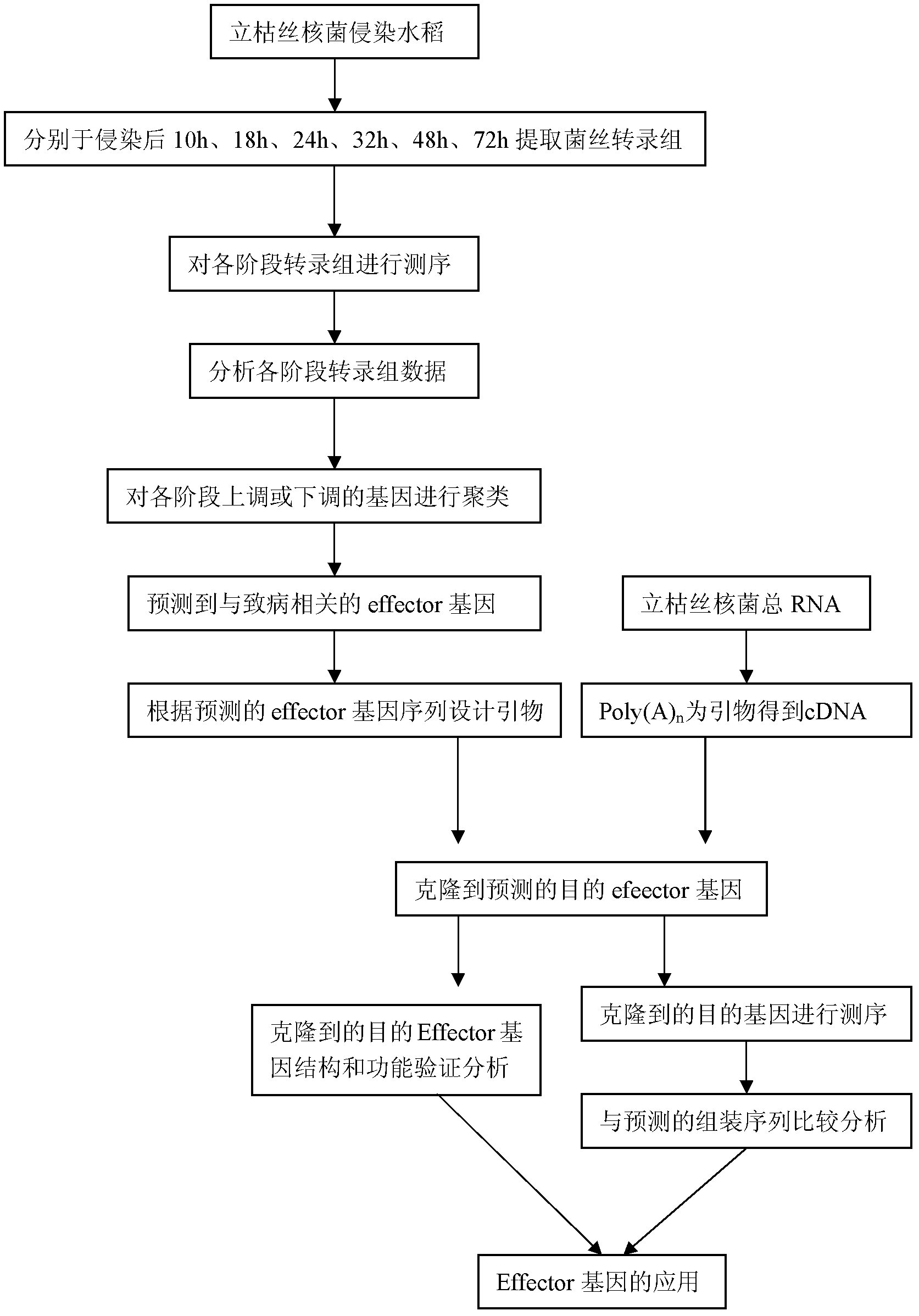
[0036] Example 1: Cloning of the predicted effector gene
[0037] The AG1IA strain was isolated and purified from rice sheath blight disease-infected plants in the field by the Rice Research Institute of Sichuan Agricultural University. It is a fungal strain that is ubiquitous on rice plants and is widely distributed in rice cultivation areas. (AG1IA strain has been published in Sichuan Agricultural University, Master's thesis, Analysis of Rhizoctonia solani AG1IA-induced corn differential protein, author Li Yun)
[0038] Under the ultra-clean workbench, use tweezers to pick out a small amount of AG1IA hyphae, insert them into 50ml PDA medium, mash the hyphae, culture at 28°C, 200r / m for two days, filter and collect the hyphae with four layers of gauze. Triturate with liquid nitrogen to extract total RNA. Use oligodT as primer and reverse transcription to get cDNA. The primers are designed according to the predicted sequence as follows:
[0039] 5’ATGCGCGGATTTACATCGTACCTTGTTGCAGCA...
Embodiment 2
[0042] Example 2: Construction, transformation and expression of prokaryotic expression vector of effector gene
[0043] The target gene obtained by PCR has no miscellaneous bands detected by electrophoresis. You can connect the target gene to the expression vector pEASY-E1 according to the instructions of TransGenBiotech Peasy-E1kit. After sequencing, the gene AG1IA10434 is 57 bases longer than the predicted sequence. The base sequence is as SEQID Shown in NO.1. The transformation conditions are the same as those of conventional Escherichia coli. The positive transformants are picked, cultured at 37°C to an OD value of 0.6, then transferred to 28°C, IPTG 1mM is induced for 7 to 11 hours, and the effector protein is expressed.
Embodiment 3
[0044] Example 3: Pathogenicity detection of expressed protein
[0045] Collect the cells expressed by AG1IA10434 and ultrasonically break them to obtain crude protein. Be careful not to denature the protein during the crushing process (the crushing time is 5S, the power should not exceed 200W, and the sample is always kept under ice bath conditions during the crushing process). Select rice leaves at the three-leaf stage cultured in a greenhouse with consistent growth and place them in a sterile petri dish with two layers of filter paper to keep the filter paper moist to keep the leaves moisturizing. Use a sterilized toothpick to make a small hole in the center of the rice leaf, and cover the small hole with a three-layer sterilized 0.5cm×0.5cm filter paper. The amount of crude protein inoculated is 50 microliters. The infected leaves were placed in a 28 degree Celsius light incubator with 12 hours of light and 12 hours of darkness, and RH (sickness index) 80%. Observe and take ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com