Engineering bacteria for expressing L-lactate dehydrogenase subjected to orthogenetic evolution and application thereof
A technology of lactate dehydrogenase and directed evolution is applied in the application field of splitting racemic mandelic acid and producing R-mandelic acid and acetophenone acid, and achieves the effects of short growth cycle, low cost and obvious effect.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0050] Example 1: Construction of engineered bacteria expressing directed evolution L-lactate dehydrogenase
[0051] (1) Prepare the genomic DNA of Pseudomonas stutzeri (Pseudomonas stutzeri SDM, CCTCC No. M206010) by conventional methods. For this process, please refer to the bacterial genome in the "Guide to Refined Molecular Biology" published by Science Press. Method of quantity preparation. Using PCR reaction, the gene encoding L-iLDH was amplified from the genome of Pseudomonas stutzeri SDM.
[0052] The PCR reaction program is:
[0053] a. 95℃ pre-denaturation for 10 minutes
[0054] b. Denaturation at 95°C for 30 seconds
[0055] c. Annealing at 60℃ for 30 seconds
[0056] d. 72℃ extension for 1 minute
[0057] Repeat the process b-d 30 cycles
[0058] e. Supplementary extension for 10 minutes
[0059] The primers used are:
[0060] Upstream primer lldU: AAGCTTATGATTTCCGCCTCTACC, containing HindIII restriction site
[0061] Downstream primer lldD: CTCGAGTCAGACGTCAGCAGACGTTG, contain...
Embodiment 2
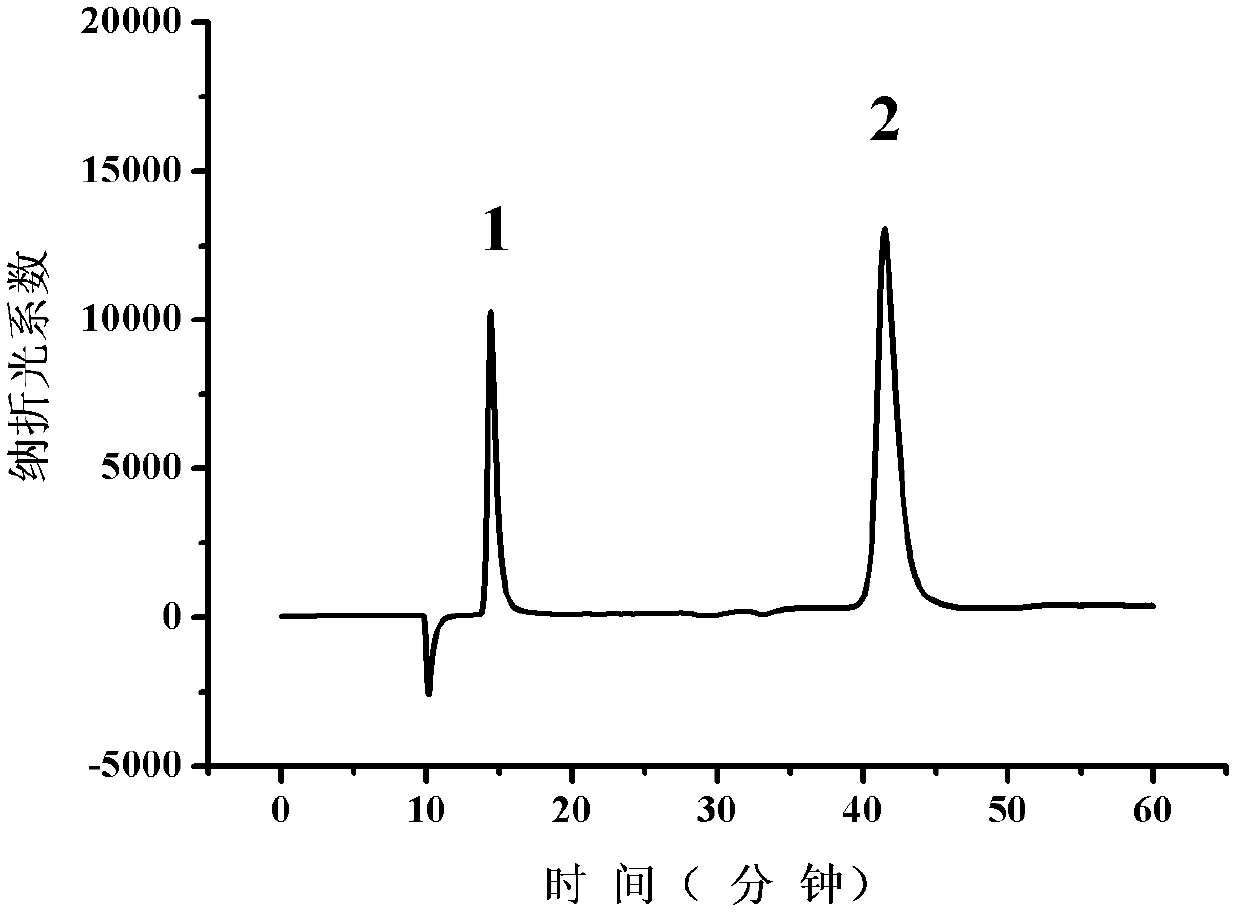
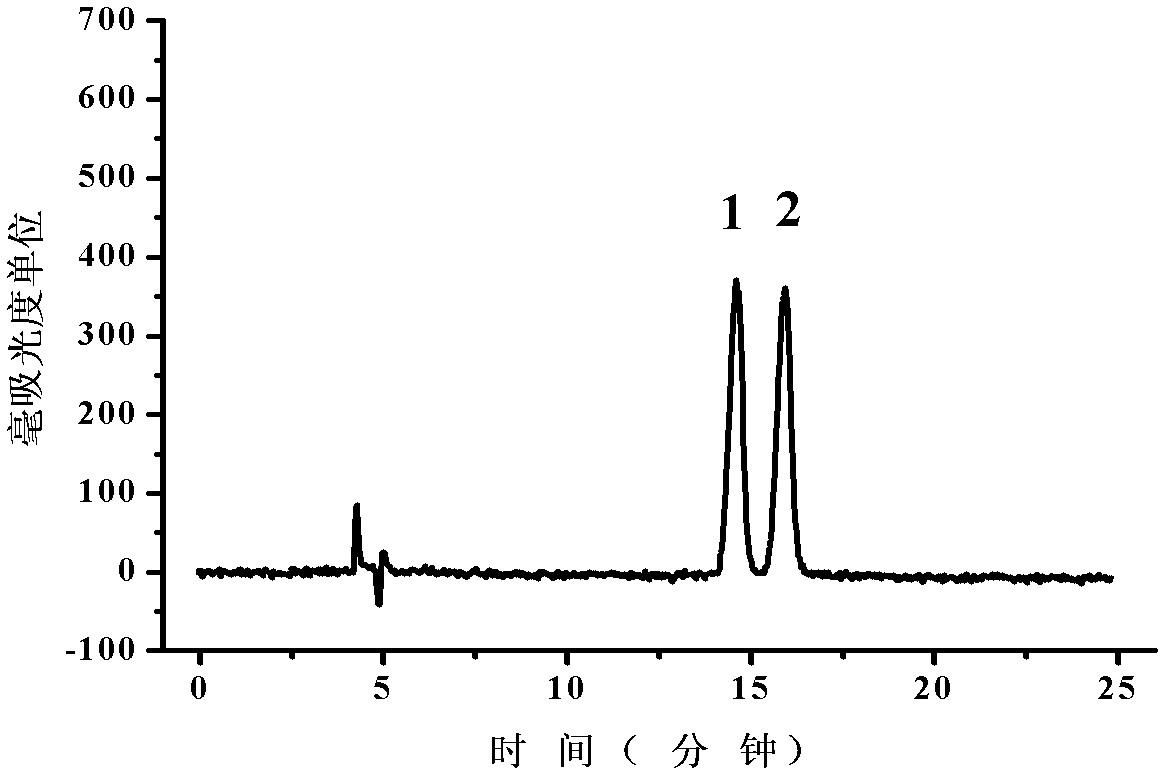
[0085] Example 2: Application of highly expressed mutant L-iLDH engineered Escherichia coli in the resolution of racemic mandelic acid
[0086] (1) Preparation of complete E. coli cells expressing mutant L-iLDH:
[0087] Plate culture: Streak the E. coli C43(DE3) strain containing pETDuet-1-mlldD vector into solid LB medium. The medium contains ampicillin at a concentration of 1 mg / ml. Cultivate for 12 hours at 37°C. Take a single colony;
[0088] Seed culture: inoculate the picked single colony into 5 ml liquid LB medium, the medium contains ampicillin at a concentration of 1 mg / ml, and culture it at 37°C for 10 hours;
[0089] Preparation of high-expressing mutant L-iLDH whole-cell catalyst: transfer the seeds to 1 liter LB medium (as for a 5-liter Erlenmeyer flask) at 1% inoculum, and cultivate at 37°C to OD 620nm When it reaches 0.6, isopropylthiogalactoside (IPTG) with a final concentration of 1 mmol / L is added to induce the expression of the mutant protein. After 6 hours of i...
Embodiment 3
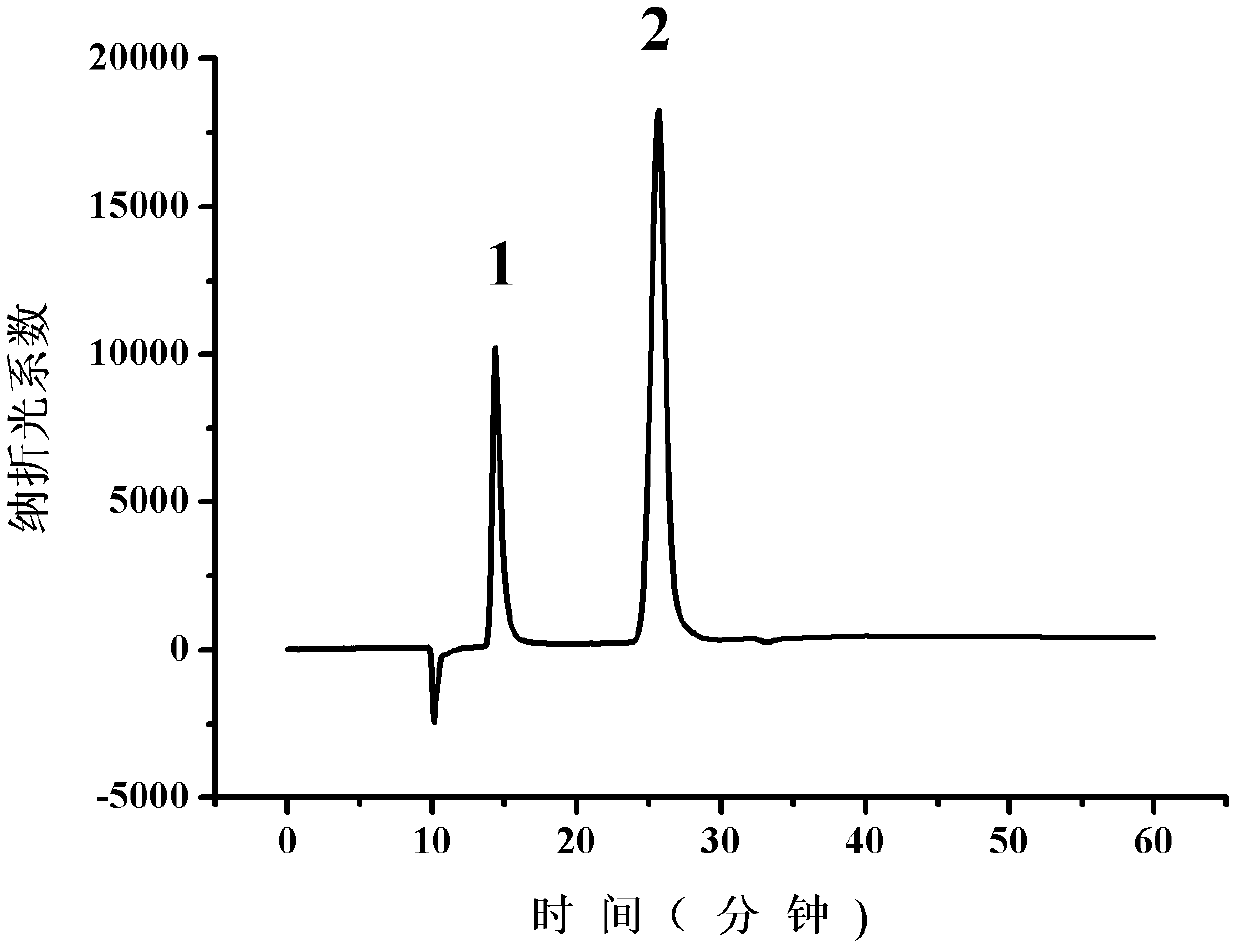
[0092] Example 3: Application of highly expressed mutant L-iLDH engineered Escherichia coli in the resolution of racemic mandelic acid
[0093] (1) Preparation of complete E. coli cells expressing mutant L-iLDH:
[0094] Plate culture: Streak the E. coli C43(DE3) strain containing pETDuet-1-mlldD vector into solid LB medium. The medium contains ampicillin at a concentration of 1 mg / ml. Cultivate for 12 hours at 37°C. Take a single colony;
[0095] Seed culture: inoculate the picked single colony into 5 ml of liquid LB medium, the medium contains ampicillin at a concentration of 1 mg / ml, cultured at 37°C for 8 hours;
[0096] Preparation of high-expressing mutant L-iLDH whole-cell catalyst: transfer the seeds to 1 liter LB medium (as for a 5-liter Erlenmeyer flask) at 1% inoculum, and cultivate at 37°C to OD 620nm When it reaches 0.5, isopropylthiogalactoside (IPTG) with a final concentration of 1.2 mmol / L is added to induce the expression of the mutant protein. After induction at 3...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com