GenomeLab eXpress Profiling (GeXP) multiplex quick detection primers and detection method for 6 food-borne pathogens
A food-borne pathogenic bacteria and detection method technology, applied in biochemical equipment and methods, microbial-based methods, microbial determination/inspection, etc., to improve sensitivity, avoid false positives and false negatives, and avoid false positives. The effect of positives and false negatives
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
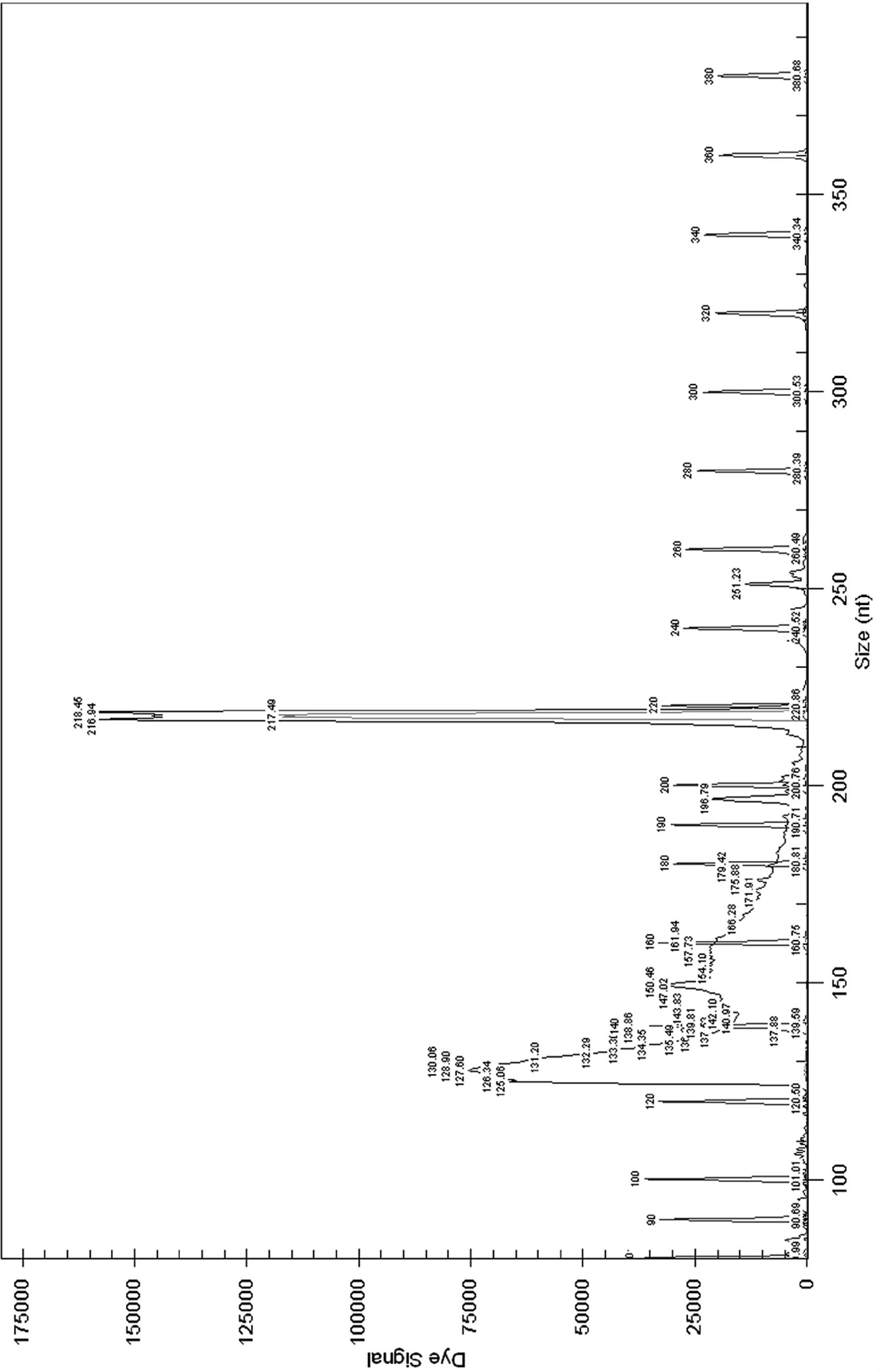
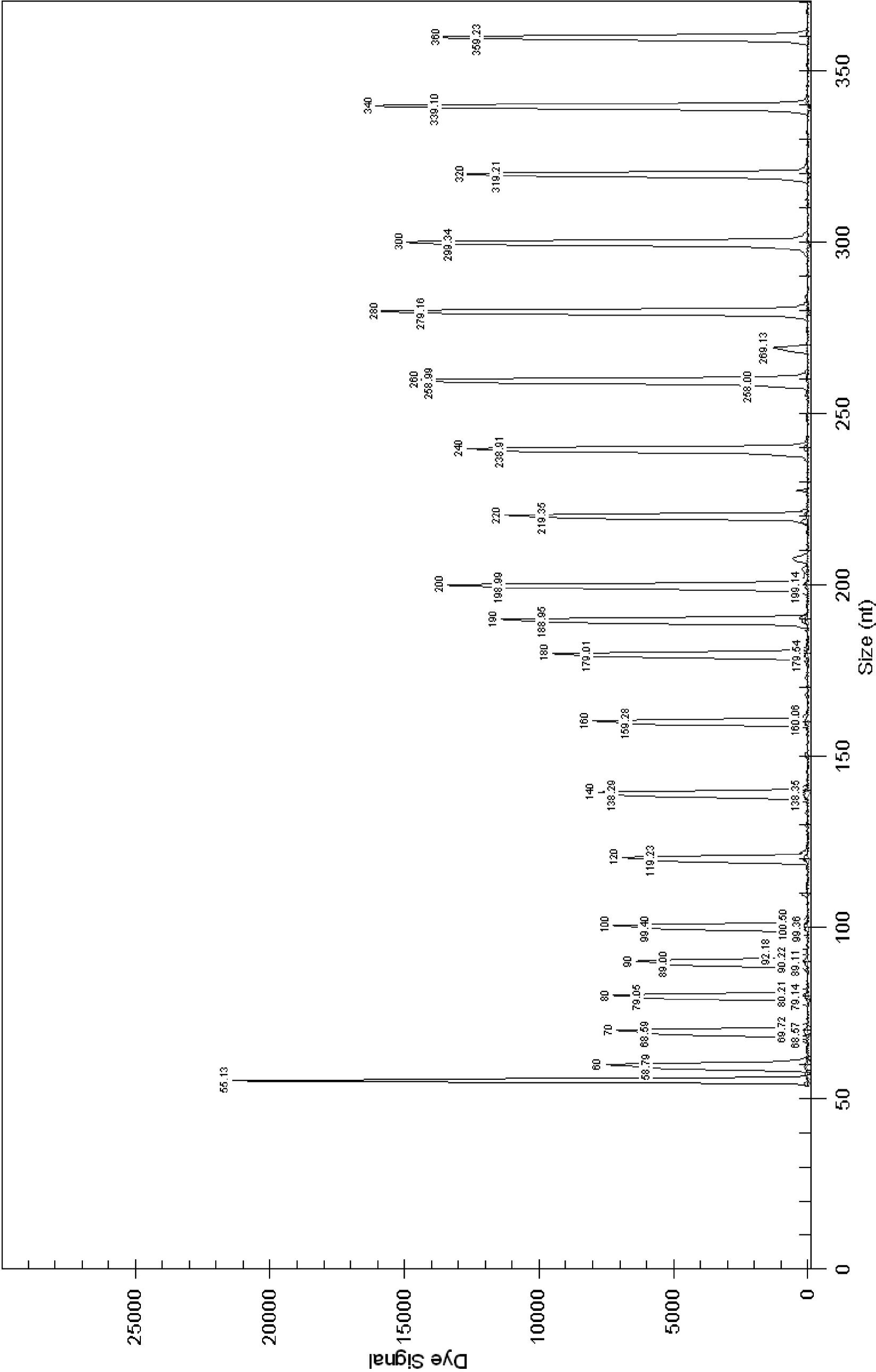
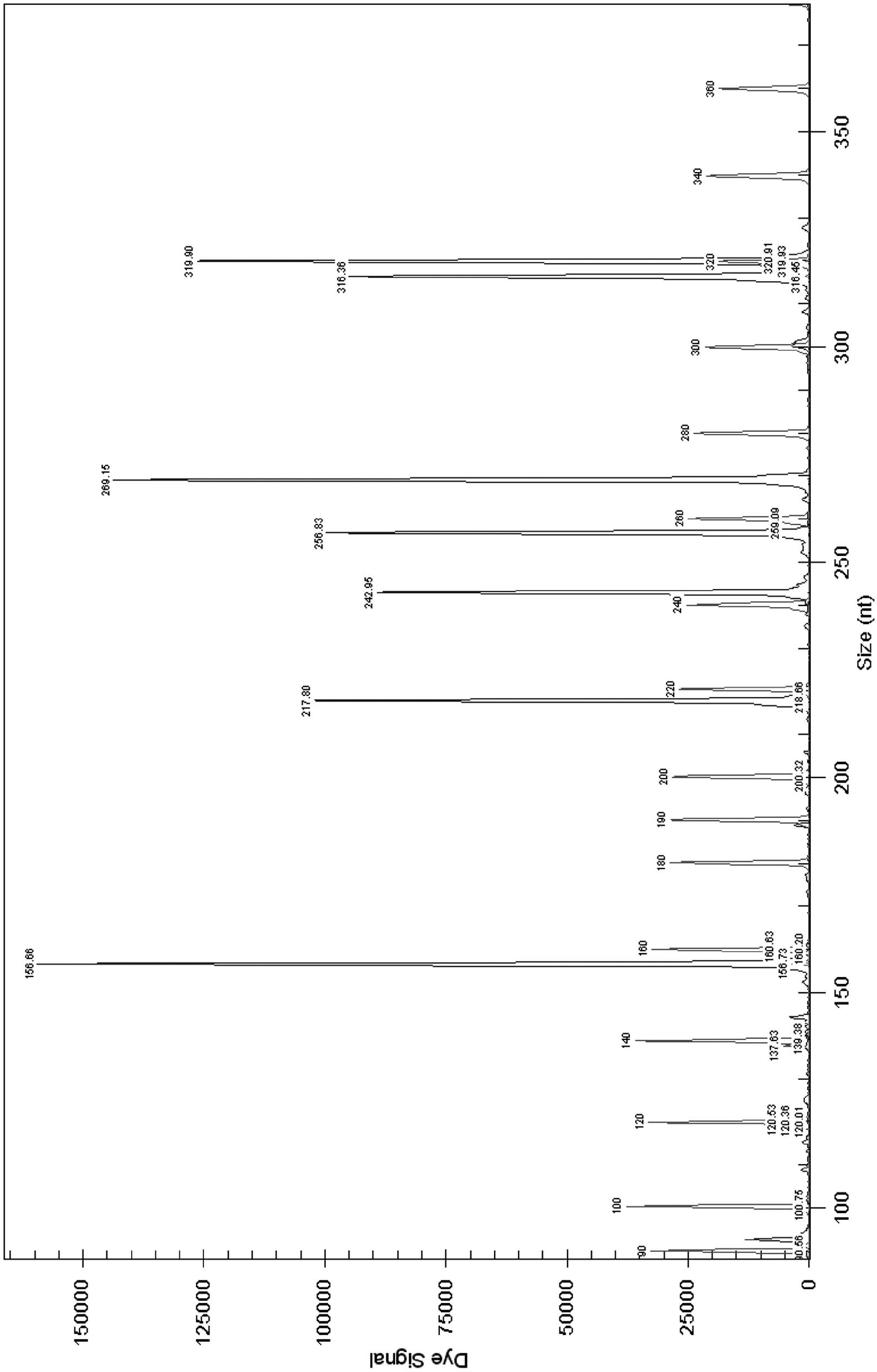
Image
Examples
Embodiment 1
[0041] Embodiment 1, the design of primer
[0042] The primers for GeXP multiplex PCR detection of 6 kinds of food-borne pathogens are a set of primers for food-borne pathogens. Each pathogenic bacteria primer set consists of two pairs of primers, one pair is specific It is a universal primer, the sequence of the universal primer is the same and the 5' end of the upstream primer is labeled with cy5 fluorescence, and each specific primer contains a sequence that can be combined with the universal primer.
[0043] 1) Design and synthesis of universal primers
[0044] The universal primer is a nucleotide coding sequence of non-biological origin, and the 5' end of the upstream primer is labeled with cy5 fluorescence. Universal primers were synthesized by Shanghai Sangon Bioengineering Technology Service Co., Ltd.
[0045] 2) Design and specificity verification of specific primers
[0046] Select DNA helicase B subunit gene (gyrB) from Salmonella, DNA helicase B subunit gene (gy...
Embodiment 2
[0055] Embodiment 2, food-borne pathogenic bacteria GeXP multiplex PCR detection method
[0056] 1) Strain culture
[0057] Staphylococcus aureus can be cultured in 10% sodium chloride tryptone soybean broth, etc., Listeria monocytogenes can be cultured in Fraser broth, etc., Campylobacter jejuni can be cultured in Brucella broth, etc. It can be cultured by nutrient broth or nutrient agar, etc. Campylobacter jejuni needs to be cultured at 42°C±1°C for 36 hours in microaerophilic conditions, and other strains need to be cultured at 37°C±1°C for 18 hours.
[0058] 2) DNA extraction
[0059] Bacterial DNA extraction commercial kit (OMEGA Bacterial DNA Kit (200)) was used to extract DNA from strain samples. The specific steps are as follows: take 1 mL of the cultured bacterial liquid and centrifuge to obtain the bacterial cells, or obtain the bacterial lawn from the nutrient agar plate. Follow the steps specified in the kit instructions. Obtain the DNA template to be tested. ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com