DNA aptamer specifically recognizing streptomycin and application of DNA aptamer
A streptomycin and aptamer technology, applied in the fields of biochemistry and molecular biology, analytical chemistry and combinatorial chemistry, can solve the problems of difficult antibody activity, long-term maintenance, high price, etc., and the method is simple, easy to operate, easy to Retouching and labeling, good stability effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
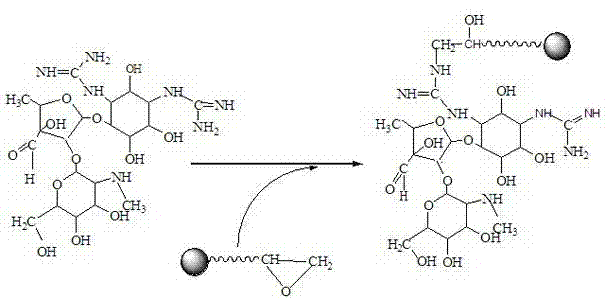
[0031]Example 1. Immobilization of streptomycin on the surface of epoxy-modified magnetic beads
[0032] The schematic diagram of the coupling of streptomycin and magnetic beads is as follows: figure 1 As shown, the specific process is:
[0033] (a) Prepare 10mM streptomycin solution, adjust the pH to 8.0, take 50μL, 10mg / mL epoxy-modified magnetic beads, wash with PBS buffer of pH 7.4 for 3-5 times, add the prepared streptomycin 200 μL of prime solution, and shake overnight at 37°C. Streptomycin not bound to the magnetic beads was washed with pH 7.4 PBS buffer and magnetically separated. Add 200 μL of 0.5 M ethanolamine at pH 8.0 to the streptomycin-magnetic bead complex, and combine with a shaker at 37°C for 6 hours to block the uncoupled groups on the magnetic beads and improve the specificity of aptamer screening. The ethanolamine not bound to the magnetic beads was washed with PBS buffer solution of pH 7.4 to obtain the affinity magnetic beads coupled with streptomyci...
Embodiment 2
[0035] Example 2. Construction of random ssDNA library and primers thereof
[0036] (a) Construction of a random ssDNA library with a length of 79 bases
[0037] 5'-TAGGGAATTCGTCGACGGATCC-N35-CTGCAGGTCGACGCATGCGCCG-3' where N represents any of the bases A, T, C, and G, and N35 represents a random fragment length of 35 bases.
[0038] (b) Synthesis of upstream primers
[0039] Upstream primer 1: 5'-TAGGGAATTCGTCGACGGAT-3'
[0040] Upstream primer 2: 5'-biotin-TAGGGAATTCGTCGACGGAT-3'
[0041] (c) Synthesis of downstream primers
[0042] Downstream primer 1: 5'-CGGCGCATGCGTCGACCTG-3'
[0043] Downstream primer 2: 5'-biotin-CGGCGCATGCGTCGACCTG-3'
Embodiment 3
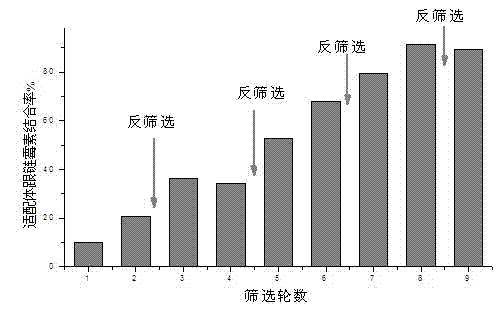
[0044] Example 3. In vitro screening of nucleic acid aptamers
[0045] In order to screen out ssDNA aptamers with high affinity and specificity to streptomycin, a total of 8 rounds of nucleic acid aptamers were screened. In order to improve the specificity of screening, a reverse screening experiment of ethanolamine-immobilized magnetic beads was carried out every two rounds of screening. In each round of screening, the binding rate of the aptamer to the target streptomycin was as follows: figure 2 shown.
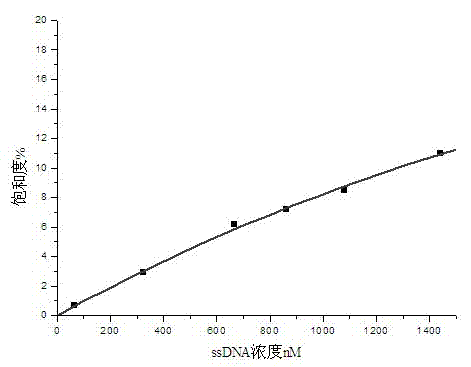
[0046] (a) Take 1 μL of ssDNA primary library, add 200 μL of binding buffer, bathe in 90°C water for 10 minutes, rapidly cool to 4°C, let stand for 15 minutes, and then stand at room temperature for 7 minutes, then add it to the affinity magnetic beads coupled with streptomycin , react at room temperature for 30min (slight shaking). Wash 5 times with binding buffer and magnetically separate to remove unbound ssDNA. Add 150 μL of elution buffer (40 mM Tris-HCl, 10 mM EDT...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com