Cell model and method for screening c-Fms kinase inhibitor
A cell and cell line technology, applied in the fields of cell biology and pharmacy, can solve the problems of cumbersome operation steps, low degree of quantification, and difficulty in accurate detection, and achieve the effect of simple, fast, and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] Example 1: Establishment of cell lines stably expressing human c-Fms and STAT1
[0060] Construct the pCORON / puro-cfms eukaryotic expression vector containing the full-length cDNA of human c-Fms (gifted by Dr. Charles Sherr and Dr. Martine Roussel, St. Jude Children's Research Hospital, USA), and liposome transfection has been fused to express GFP and STAT1 human osteosarcoma cell line.
[0061] A novel cell line stably expressing human c-Fms was obtained by resistance screening and limiting dilution. Use DMEM medium containing 4500 mg / L glucose, 10% fetal bovine serum, 4 mM L-glutamine, 500 μg / mL G418 and 2 μg / mL puromycin at 37 °C, 5% CO 2 , cultivated in an 80% humidity incubator.
[0062] Compared with before transfection, the morphology of the obtained cells did not change significantly, and the growth state was stable.
[0063] The cell line prepared in this example (ie, the preserved human osteosarcoma cell line in the present invention, with the deposit num...
Embodiment 2
[0064] Example 2: Identification of newly established cell lines
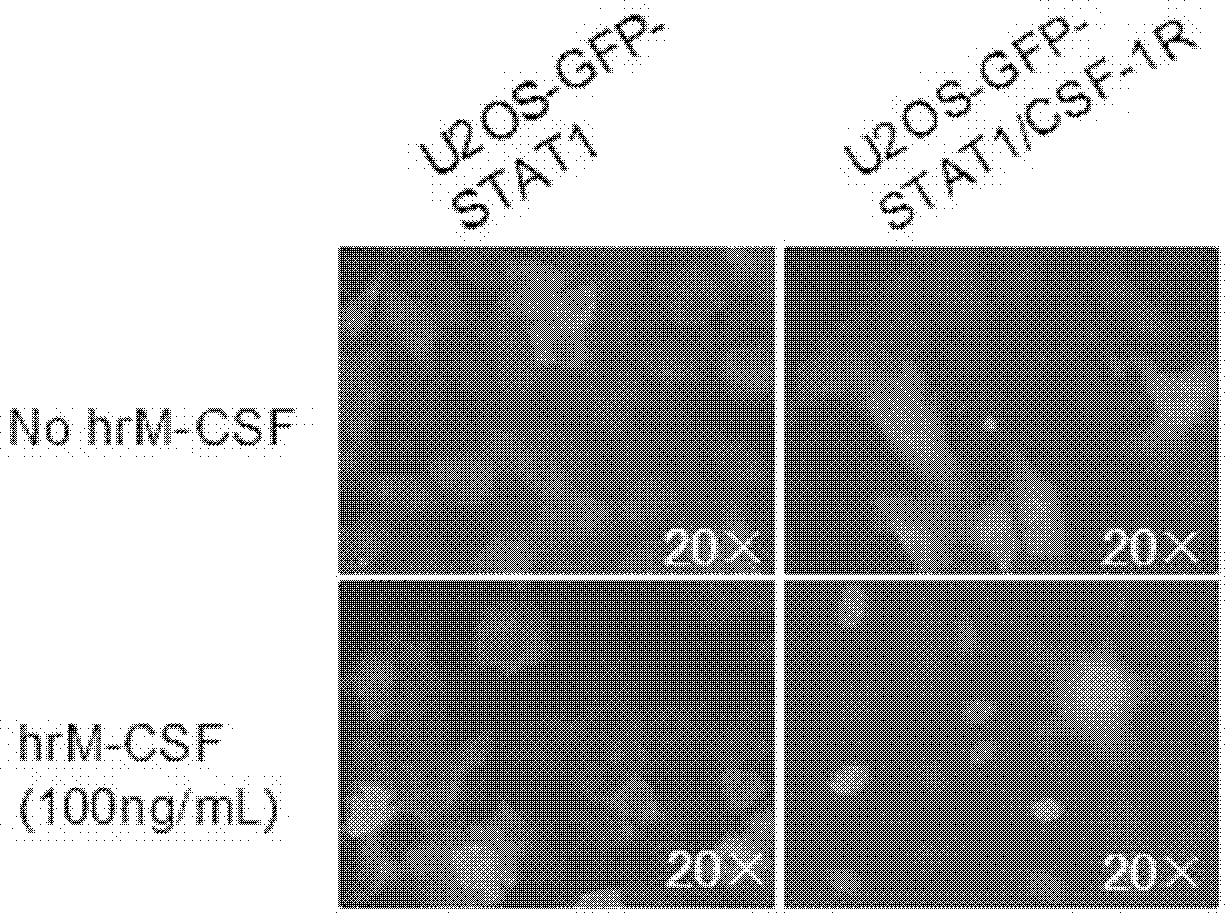
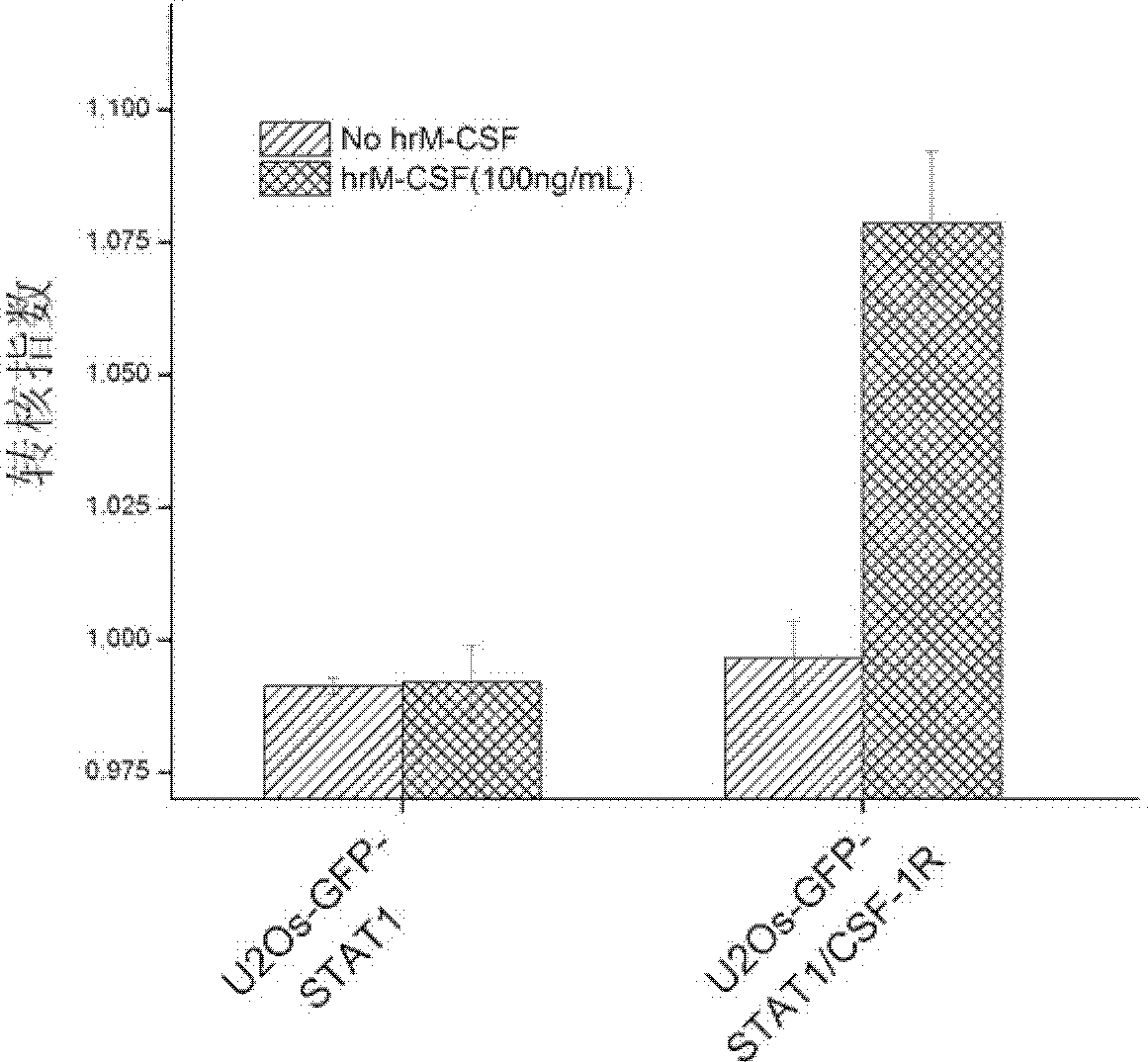
[0065] The cell line prepared in Example 1 was treated with recombinantly expressed human M-CSF (hrM-CSF) to activate c-Fms and induce nuclear translocation of GFP-STAT1. After the human macrophage colony-stimulating factor receptor in the established cell line binds to its specific ligand M-CSF, the receptor dimerizes and the spatial conformation changes to activate the kinase domain. Through transphosphorylation, the signaling pathway downstream of c-Fms is activated. Phosphorylated GFP-STAT1 also forms dimerization, enters the nucleus, and undergoes nuclear translocation.
[0066] In order to quantitatively detect the degree of nuclear translocation of GFP-STAT1 induced by hrM-CSF, in this example, In Cell Analyzer 1000 or In Cell Analyzer 2000 was used to obtain cell images of GFP-STAT1 nuclear translocation, which were analyzed by the Nuclear Trafficking Analysis Module. The cell image of , the degree ...
Embodiment 3
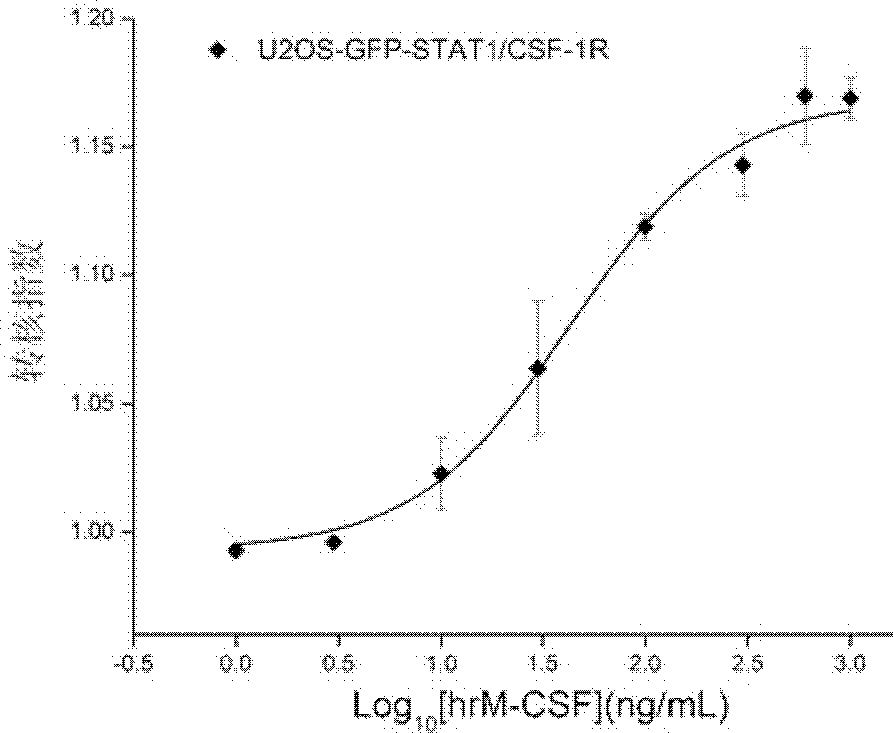
[0074] Example 3: Determination of the half effective concentration of hrM-CSF-induced GFP-STAT1 nuclear transfer
[0075] Proceed as follows:
[0076] 1) The cells prepared in Example 1 were made into 1×10 5 cells / mL of cell suspension, inoculated in a black transparent bottom 96-well culture plate, 100 μL / well.
[0077] 2) At 37°C, 5% CO 2 , cultivated in an 80% humidity incubator for 24 hours.
[0078] 3) Wash the cells twice with the cell analysis solution, 100 μL / well each time; discard the solution, and then add 50 μL / well of the cell analysis solution.
[0079] 4) Add hrM-CSF diluted in the cell analysis solution to make the final concentrations respectively 1, 3, 10, 30, 100, 300, 600, and 1000 ng / mL.
[0080] 5) 37°C, 5% CO 2 After incubating in an 80% humidity incubator for 30 minutes, add 37°C preheated cell fixation solution, 100 μL / well, shake the culture plate slightly to mix, and place it at room temperature in the dark for 20 minutes.
[0081]6) Discard...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com