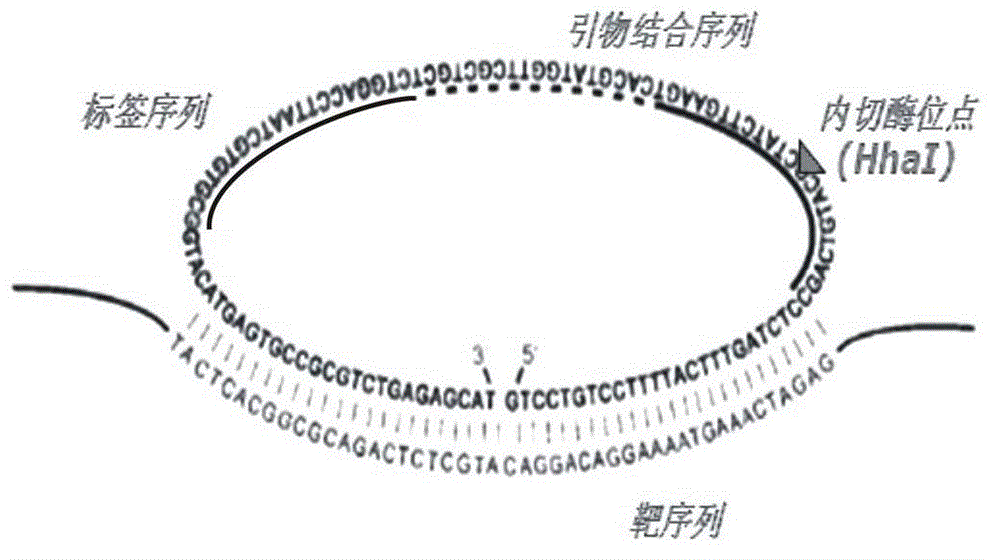
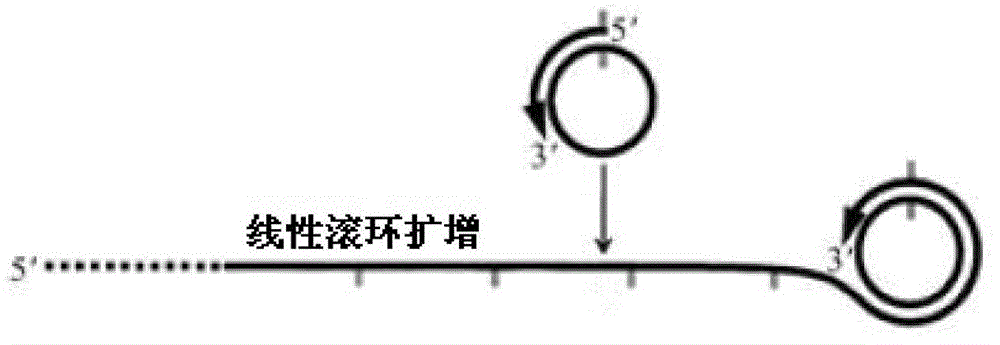
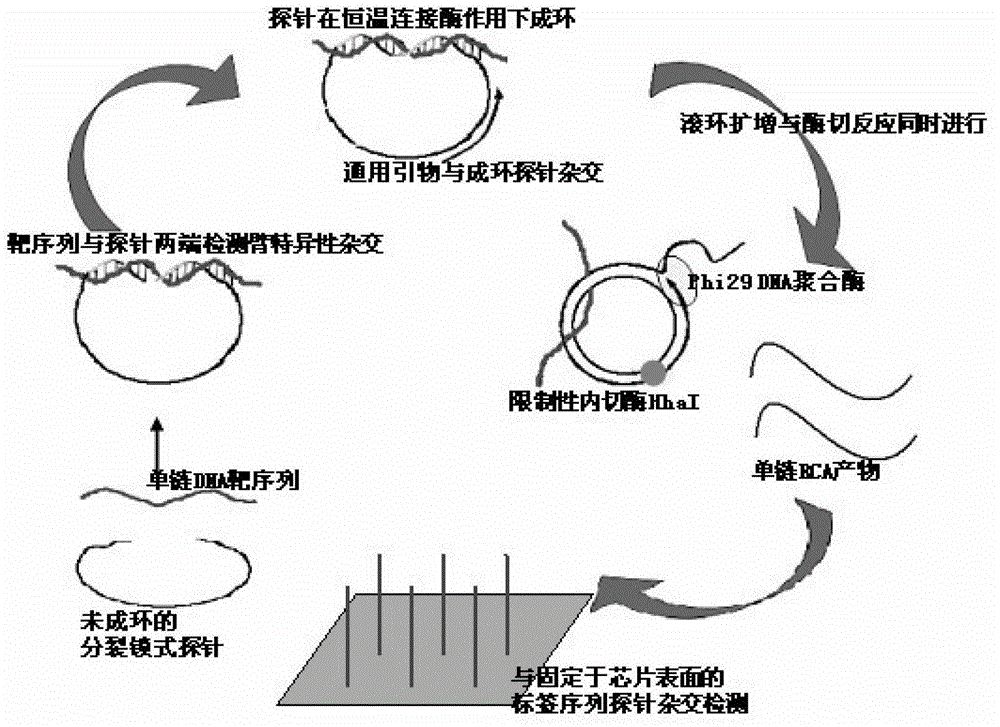
Multi-RCA (rolling circle amplification) method based on split padlock probes
A padlock probe and multiplex technology, applied in the field of multiplex RCA based on split padlock probes, can solve the problems of high false negative, high cost, complicated steps, etc., and achieve high specificity, convenient operation and strong specificity. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] Example 1: Taking isoniazid-resistant clinical isolates of Mycobacterium tuberculosis (katG315 mutation) as an example of the target sequence probe, the specific operation steps are as follows:
[0046] Step 1. Ligation reaction: take 1uL of the genome extract of clinical isolates and 0.1μmol / L split padlock probe (the final concentration of the four probes is 0.1umol / L), the base sequence of which is shown in the sequence table sequence 11, the target sequence Mix with the probe, boil and denature, immediately place it on ice for 5 minutes, take it out, raise the temperature to 37°C, hybridize for 15 minutes, add 2.5U of T4 DNA ligase and 1 μL of T4 DNA ligase buffer, deionized water to make up 10uL of the reaction system, and react The time is 45min, then add 10U exonucleaseI and 10U exonucleaseIII, and react at 37°C for 15min to prepare a ligation product containing a circular template; the composition of the T4DNA ligase buffer is 40mmol / L Tris-HCL, 10mmol / L MgCL 2 ...
Embodiment 2
[0055] Example 2: Taking isoniazid-resistant clinical isolates of Mycobacterium tuberculosis (inhA-15 mutation) as an example of the target sequence probe, the specific operation steps are as follows:
[0056] Step 1. Ligation reaction: Take 1uL of the genome extract of the clinical isolate and 0.1μmol / L split padlock probe (the final concentration of the four probes is 0.1umol / L). Mix with the probe, boil and denature, immediately place it on ice for 5 minutes, take it out, raise the temperature to 37°C, hybridize for 15 minutes, add 2.5U of T4 DNA ligase and 1 μL of T4 DNA ligase buffer, deionized water to make up 10uL of the reaction system, and react The time is 45min, then add 10U exonucleaseI and 10U exonucleaseIII, and react at 37°C for 15min to prepare a ligation product containing a circular template; the composition of the T4DNA ligase buffer is 40mmol / L Tris-HCL, 10mmol / L MgCL 2 ,10mmol / L DTT and 0.5mmol / L ATP;
[0057] Step 2. Rolling circle amplification reaction...
Embodiment 3
[0060] Example 3: Taking rifampicin (RFP) drug-resistant clinical isolates (rpoB531 mutation) of Mycobacterium tuberculosis as an example of the target sequence probe, the specific operation steps are as follows:
[0061]Step 1. Ligation reaction: Take 1uL of the genome extract of the clinical isolate and 0.1μmol / L split padlock probe (the final concentration of the four probes is 0.1umol / L). Mix with the probe, boil and denature, immediately place it on ice for 5 minutes, take it out, raise the temperature to 37°C, hybridize for 15 minutes, add 2.5U of T4 DNA ligase and 1 μL of T4 DNA ligase buffer, deionized water to make up 10uL of the reaction system, and react The time is 45min, then add 10U exonucleaseI and 10U exonucleaseIII, and react at 37°C for 15min to prepare a ligation product containing a circular template; the composition of the T4DNA ligase buffer is 40mmol / L Tris-HCL, 10mmol / L MgCL 2 ,10mmol / L DTT and 0.5mmol / L ATP;
[0062] Step 2. Rolling circle amplificati...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com