Method for testing and identifying cepaea hortensis through polymerase chain reaction (PCR)
A snail and forest technology, applied in the field of molecular biology detection and identification, can solve problems such as difficult to distinguish, difficult to identify eggs and young snails, little knowledge about the classification of terrestrial molluscs, etc., to achieve good practicability, rapid and reliable detection and Strong identification and practical effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
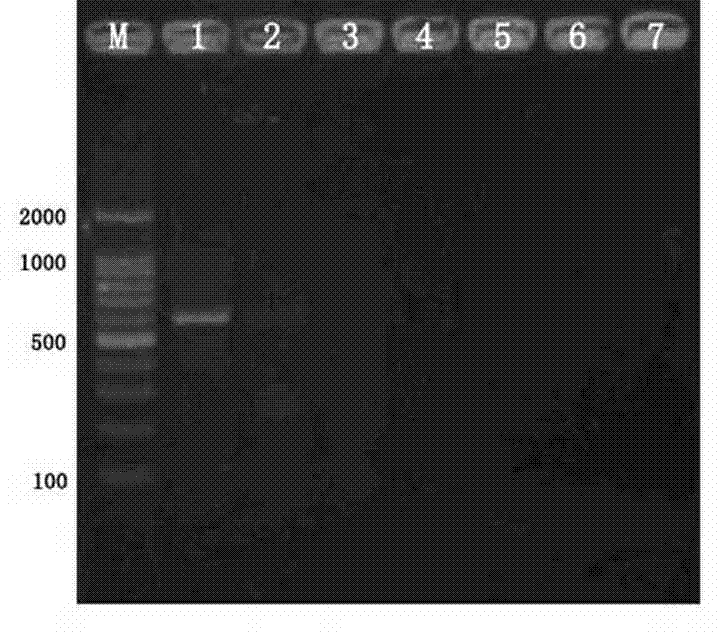
[0016] Example 1: Forest onion snail PCR primer specific test
[0017] 1. Preparation of materials
[0018] Test snails are as follows: Forest onion snails Cepaea Nemoralis Garden onion snail Cepaea hortensis ; Sanda snail Helix aspersa ; Cover the big snail Helix pomatia ; Sven Hao's big snail NESIOHELIX SWINHOEI ; Bradybaena (( Acusta Cure ravida ravida ; Mediterranean white snail Cernuella Virgata Essence
[0019] The above-mentioned trial snails are intercepted in the nationwide port quarantine. After confirming the key laboratory of the State Inspection and Quarantine of the State Inspection and Quarantine of the Quality Inspection and Quarantine, it is stored at -20 ° C for later use.
[0020] 2. The establishment of the PCR method
[0021] 2.1, the design synthesis of primers: based on forest -based onion snail CO i (mitochondrial cytochromes C oxidase sub -meal I) gene sequence, design and analyze primers with primers of PRIMER 5 and Oligo 6, and submit it after testing it...
Embodiment 2
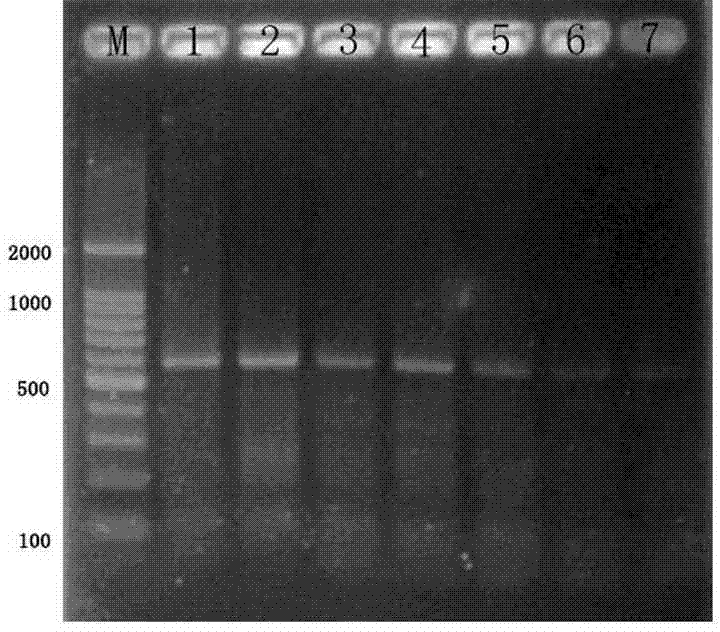
[0032] Example 2: Forest onion snail PCR test sensitivity test
[0033] Use the 10x concentration series to dilute method to dilute the forest snail DNA raw liquid (100NG / μL) extracted from the Example 1 to 10ng / μL, 1ng / μL, 100 PG / μL, 10 PG / μL, 1 PG / μL, 100FG / μL and 10 FG / μL have 7 different concentration gradients.
[0034] PCR amplification reaction system: The overall accumulation is 25 μL, of which 2 × TAQ PCR MasterMIX mixed solution is 12.5 μl, the upstream and downstream primers are 0.5 μl, the DNA template is 2 μL, and the remaining sterilizer DDH 2O complement.Put it in the PCR amplifier for amplification after mixing.
[0035] The PCR amplification reaction program is: 95 ° C premium 5min; 95 ° C 50s, 52 ° C 30s, 72 ° C 50s, 35 cycles; 72 ° C extension for 10 minutes, ending the reaction.
[0036] PCR product testing and identification: Take PCR products 10 μL at 1.5%agarose gel (containing bromide ingot) with 98V electrophoresis 40min, and observe with gel imaging instr...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com