Engineering bacterium and application thereof in production of amorpha fruticosa-4,11-diene
A technology of engineering bacteria and amorpha, applied in the direction of microorganism-based methods, bacteria, microorganisms, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0070] Embodiment 1, the preparation of DNA molecule
[0071] 1. Prepare Amorpha-4,11-diene synthase gene (ADS gene, according to the nucleotide sequence of the ADS gene in Artemisia annua, the DNA sequence was optimized according to the preference of Escherichia coli)
[0072] A double-stranded DNA molecule (1672bp) shown in Sequence 1 of the Sequence Listing was synthesized.
[0073] In Sequence 1 of the sequence listing, the 1st to 6th nucleotides from the 5' end are the restriction endonuclease XhoI restriction endonuclease recognition sequence, the 23rd to 1660th nucleotides are the coding region of the ADS gene, and the 1661st to 1660th nucleotides are the coding region of the ADS gene. Nucleotide 1666 is two consecutive stop codons, and nucleotides 1667 to 1672 are the restriction endonuclease BamHI recognition sequence.
[0074] 2. Preparation of HMG-CoA reductase gene (mvaE gene, biological source is Enterococcus faecalis)
[0075] A double-stranded DNA molecule (24...
Embodiment 2
[0101] Embodiment 2, construction of plasmid
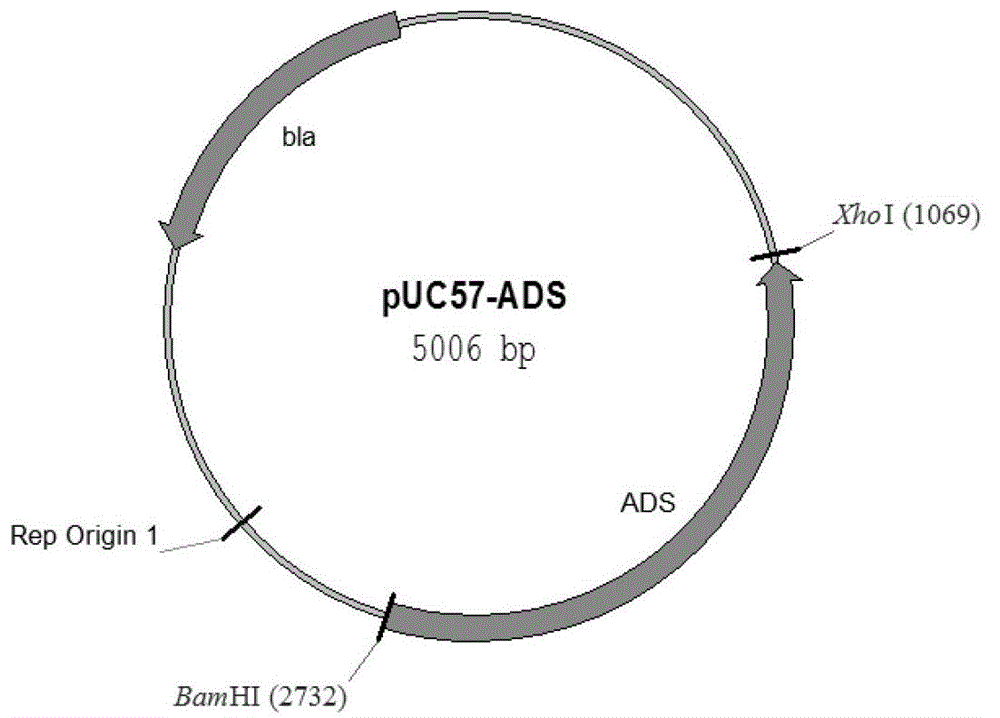
[0102] 1. Construction of plasmid pUC57-ADS
[0103] 1. Digest the double-stranded DNA molecule shown in Sequence 1 of the Sequence Listing with restriction endonucleases Xho I and BamHI, and recover the digested product.
[0104] 2. Digest vector pUC57 with restriction endonucleases Xho I and BamHI to recover a vector backbone of about 2700 bp.
[0105] 3. Ligate the digested product of step 1 with the vector backbone of step 2 to obtain the recombinant plasmid pUC57ADS (see figure 1 ).
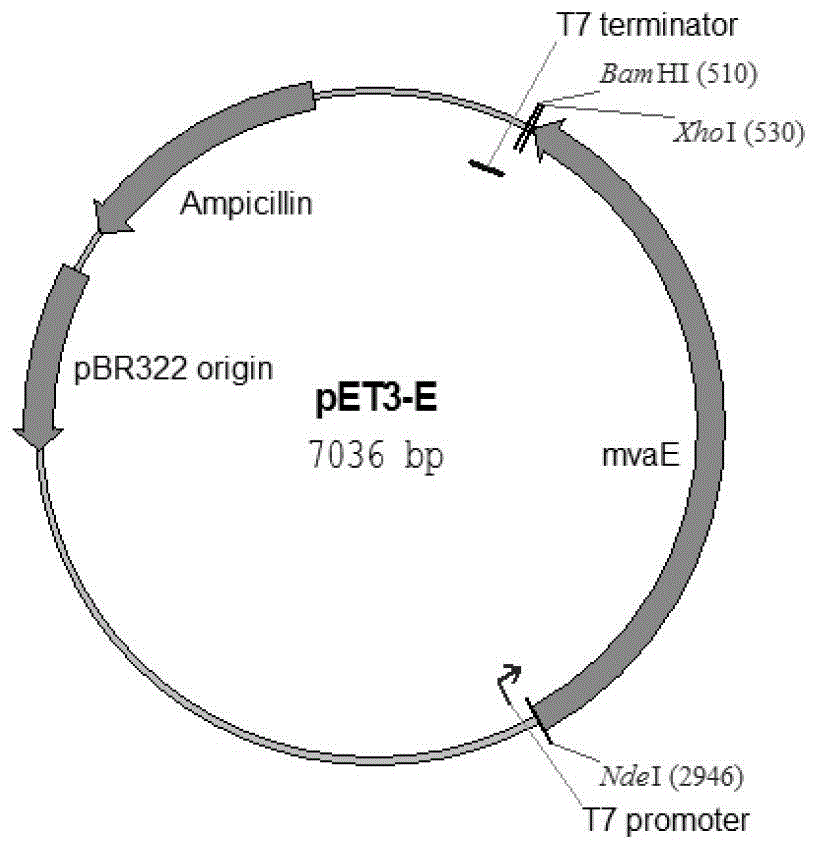
[0106] 2. Construction of plasmid pET3-E
[0107] 1. Digest the double-stranded DNA molecule shown in Sequence 2 of the Sequence Listing with restriction endonucleases Nde I and BamH I, and recover the digested product.
[0108] 2. Digest the vector pET3b with restriction endonucleases Nde I and BamH I to recover a vector backbone of about 4600 bp.
[0109] 3. Ligate the digested product of step 1 with the vector backbone of step 2 to obtai...
Embodiment 3
[0183] Embodiment 3, the construction of recombinant bacteria
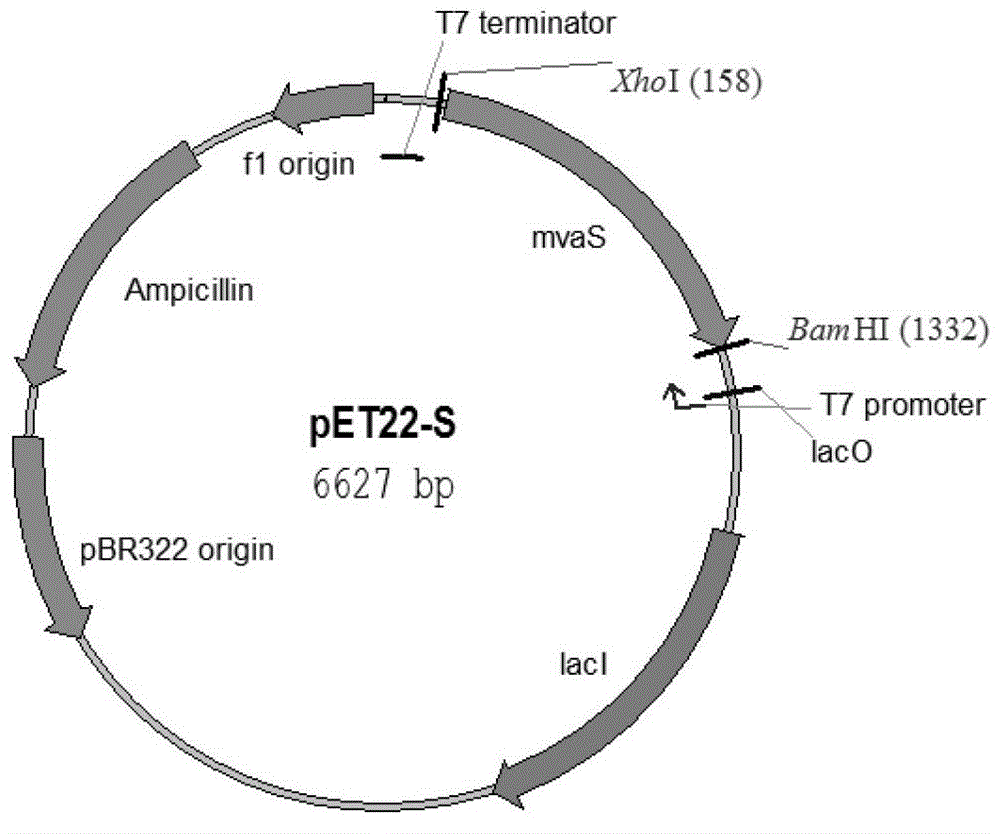
[0184] 1. Construction of recombinant bacteria pET3-ES / DHGT7
[0185] The recombinant plasmid pET3-ES was introduced into Escherichia coli DHGT7 competent cells to obtain the recombinant bacteria, which was named recombinant bacteria pET3-ES / DHGT7 (produced mevalonic acid).
[0186] 2. Construction of recombinant bacteria pET3-ES / pAOC3-MD-ispA-ADS / DHGT7
[0187] The recombinant plasmid pET3-ES and the recombinant plasmid pAOC3-MD-ispA-ADS were jointly introduced into Escherichia coli DHGT7 competent cells to obtain recombinant bacteria, which were named recombinant bacteria pET3-ES / pAOC3-MD-ispA-ADS / DHGT7 (producing Amorpha-4,11-diene).
[0188] 3. Construction of recombinant bacteria DHGT7ES
[0189] 1. The plasmid pKDS (for the schematic diagram of the structure, see Figure 16 ) into Escherichia coli DHGT7 competent cells to obtain recombinant bacteria, which were named as recombinant bacteria pKDS / DHGT7. ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com