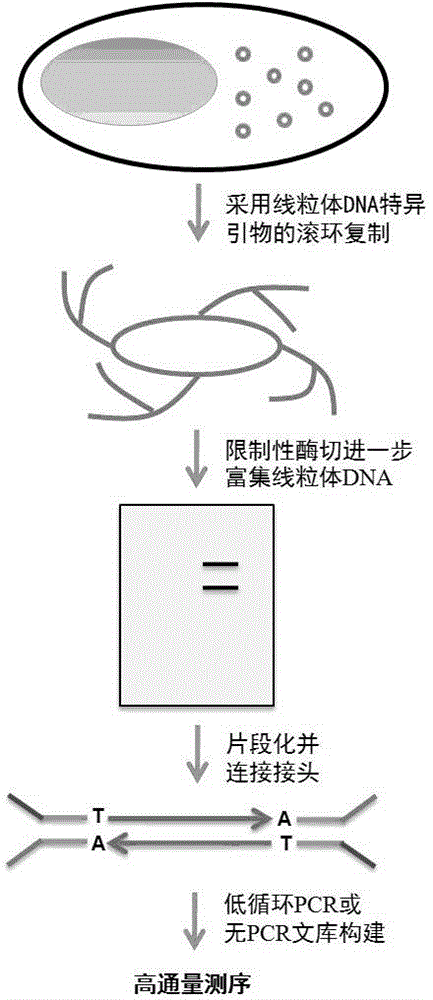
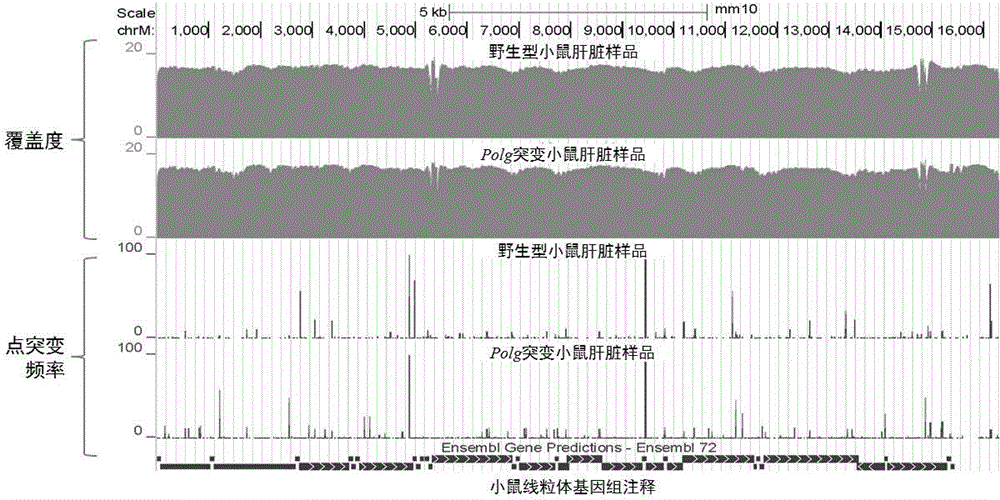
Method for precisely determining high-frequency and low-frequency mutations of mitochondrial DNA (deoxyribonucleic acid) by high-throughput sequencing
A low-frequency mutation and mitochondrial technology, applied in the field of genomics, can solve the problems of poor uniformity of mitochondrial genome coverage, time-consuming and labor-intensive problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0022] Embodiment 1: the extraction of total DNA
[0023] Total DNA (including human, orangutan, macaque, rat, mouse, Drosophila, zebrafish, nematode, Arabidopsis, rice, cotton, rapeseed, yeast and Plasmodium) was extracted using Qiagen’s QIAamp DNA Blood Mini kit Reagent test kit. Different cells or tissues were extracted according to the procedures suggested by Qiagen. Typically, 20 mg of tissue is sampled and 200 μl of AL buffer is added. Homogenization was carried out with steel balls in a 2ml round bottom centrifuge tube. The instrument used was a TissueLyser homogenizer from Qiagen Company with a parameter of 50 beats per second for a total of 2 minutes of homogenization. After column purification, elute into 200 μl of pure water. DNA concentration was determined using a Nanodrop spectrophotometer. 100 ng of total DNA was used to construct the mitoRCA-seq library.
Embodiment 2
[0024] Example 2: Mitochondrial DNA Amplification Using Rolling Circle Replication
[0025] 100 ng of total DNA was used for rolling circle amplification. Add 100ng of total DNA to a 50ul reaction system, which also includes: 1x Phi29 DNA polymerase buffer (NEB), 0.2 μg / ml BSA, 1 mM dNTP (NEB) and 25 μM mitochondrial DNA-specific primer (mouse See the sequence listing for primers). The system was mixed and heated at 95°C for 3 minutes, then cooled to room temperature, followed by the addition of 1 μl Phi29 DNA polymerase (10 unit / μl, NEB). After mixing, react at 37°C for 16 hours. This was followed by heating to 65°C for 10 minutes to inactivate the polymerase. Qiagen's REPLI-g Mitochondrial DNA kit can also be used for rolling circle replication.
Embodiment 3
[0026] Example 3: Restriction Enzyme Digestion
[0027] Different species require different restriction enzymes. The principle is to choose 1-2 restriction endonucleases that can make the corresponding mitochondrial DNA produce 2 bands that can be separated on agarose gel electrophoresis. We chose EcoRV (NEB) to generate 2 bands of 9.5kb and 6.8kb. For Drosophila, NdeI (NEB) and EcoRV (NEB) can be used to generate 10kb and 9.8kb bands. For human samples, SacI(NEB) can be used to generate 2 bands at 6.9kb and 9.6kb. The specific restriction enzyme digestion system is implemented according to the recommended dosage of each enzyme.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com