Real-time fluorescent PCR (polymerase chain reaction) kit and oligonucleotides for identifying zygosaccharomyces rouxii
An oligonucleotide and real-time fluorescence technology, applied in the field of inspection and quarantine, can solve the problems of cumbersome operation, fast customs clearance at unfavorable ports, long identification cycle, etc., to ensure food safety, optimize reaction conditions, and have good specificity and stability. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Design and synthesis of embodiment 1 primers and probes
[0041] In the present invention, firstly, sequence comparison and analysis are carried out on Luxie conjugative yeast and its nine similar yeasts, and the ITS1, 5.8S, and ITS2 gene sequences of these more than ten kinds of yeasts are used to establish a local database. ITS1, 5.8S, and ITS2 gene sequences were extracted using Python language scripts, and independent databases of ITS1, 5.8S, and ITS2 genes were established for ten yeast species. Use MEGA4 for sequence alignment, and use Python language scripts to find the conserved sequences in different types of yeast and the variant sequences among different types of yeast. Species-specific primers and probes were designed using Becaon software considering conserved sequences and variant sequences. Perform electronic PCR according to the obtained primers and probes to obtain the theoretical amplified sequence, and on this basis, perform BLAST on the theoreticall...
Embodiment 2
[0046] Embodiment 2 identifies the real-time fluorescent PCR kit of Ruby's binding yeast
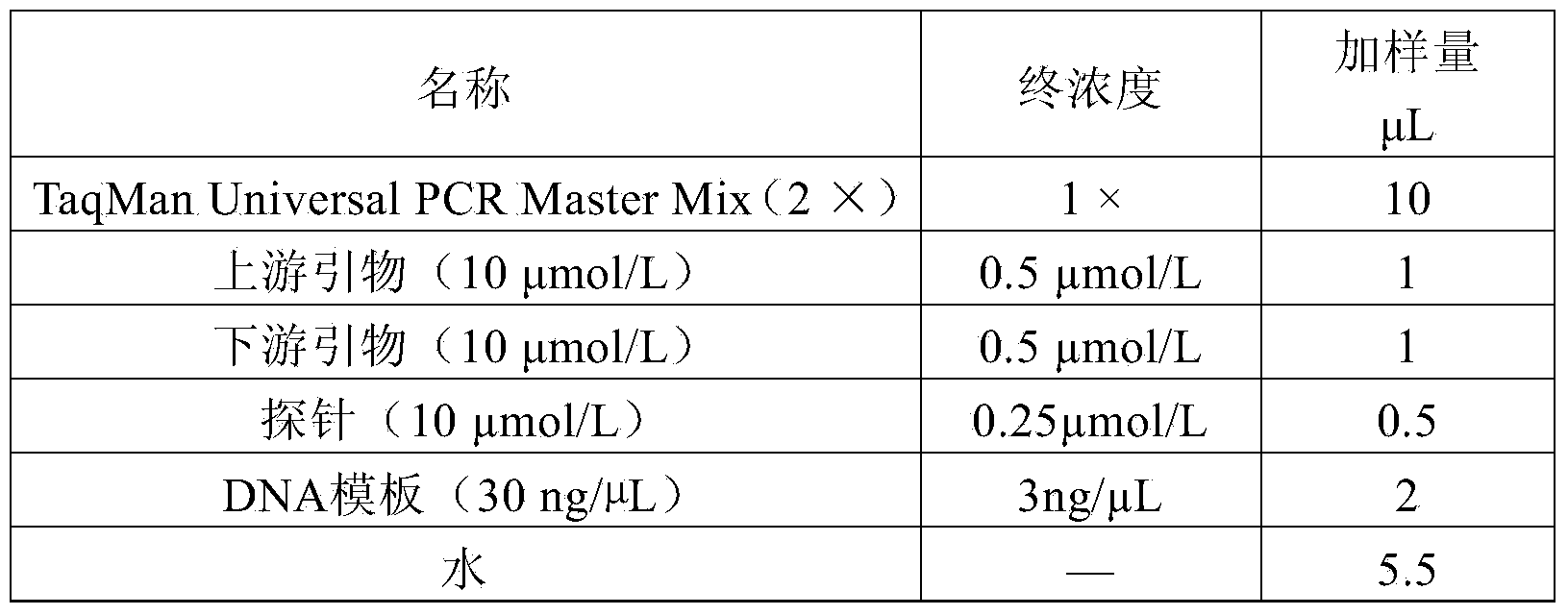
[0047] (1) Real-time fluorescent PCR reaction solution, which includes the upstream primer shown in SEQ ID No.1 in the sequence table, the downstream primer shown in SEQ ID No.2 in the sequence table, and the fluorescent probe shown in SEQ ID No.3 in the sequence table , the 5' end of the probe is labeled with the reporter fluorophore FAM, and the 3' end is labeled with the quencher fluorophore BHQ1;
[0048] (2) Negative control: sterile double distilled water;
[0049] (3) Positive control: Zygosaccharomyces rouxii CGMCC2.1915.
[0050] Further, the real-time fluorescent PCR reaction solution also includes reagents and enzymes required for conventional real-time fluorescent PCR reactions. Preferably, the commercially purchased TaqMan Universal PCR Master Mix (2×) used in the present invention is purchased from Beijing Dongfang Jiacheng Science and Trade Co., Ltd.
Embodiment 3
[0051] Embodiment 3 specificity test
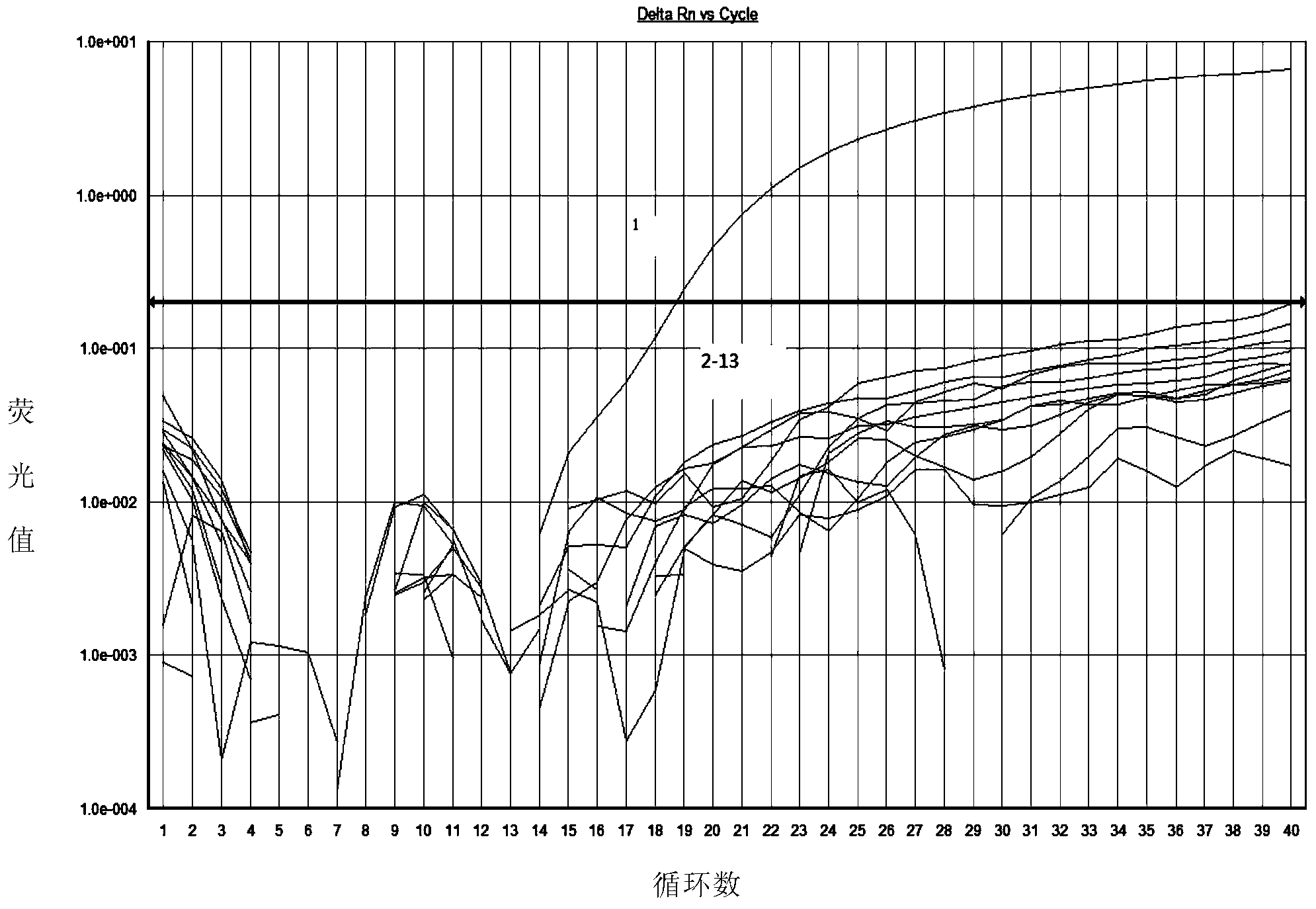
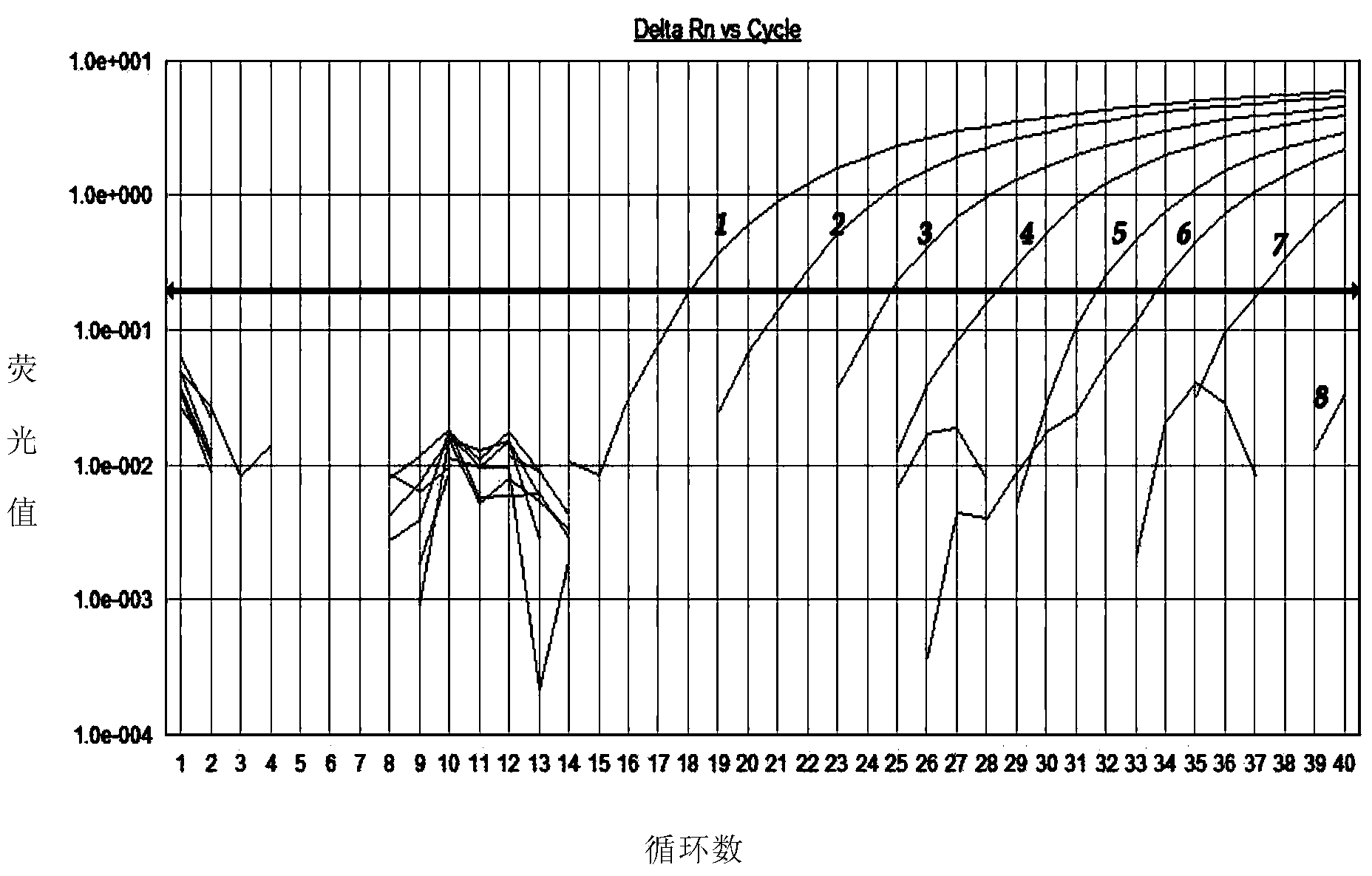
[0052] The present invention selects Saccharomyces cerevisiae CGMCC2.1882, Candida parasilosis ATCC22019, Pichia carsonii CGMCC2.1908, Debaryomyces hansenii CGMCC2.1595 , Dekkera bruxellensis JCM11407, Rhodotorula glutinis CGMCC2.0703, Pichia membranifaciens CGMCC2.1705, Bayer combined yeast Zygosaccharomyces bailii (CGMCC2.1452), Dispora combined yeast (Zygosaccharomyces bisporus) CGMCC2.1529, Escherichia coli (E.coli) ATCC25922, Staphylococcus aureus (Staphylococcus aureus) CMCC26001 were used as samples to be tested. ), the Japanese Culture Collection (JCM) and the American Type Culture Collection (ATCC). Zygosaccharomyces rouxii CGMCC2.1915 was used as a positive control.
[0053] The genomic DNA of the above-mentioned strains to be tested and the positive control were extracted respectively, the DNA concentration was adjusted to 30 ng / μL, and real-time fluorescent PCR amplification reactions were carried out according to the follow...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com