Molecular inversion probe for detecting YVDD drug-resistant mutation of hepatitis B virus and application of molecular inversion probe
A molecular inversion probe, hepatitis B virus technology, applied in microorganism-based methods, recombinant DNA technology, microbial determination/inspection, etc., can solve the problems of complex DNA sequencing operations, difficult to standardize results, poor repeatability, etc. Achieve low cost, high specificity, and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
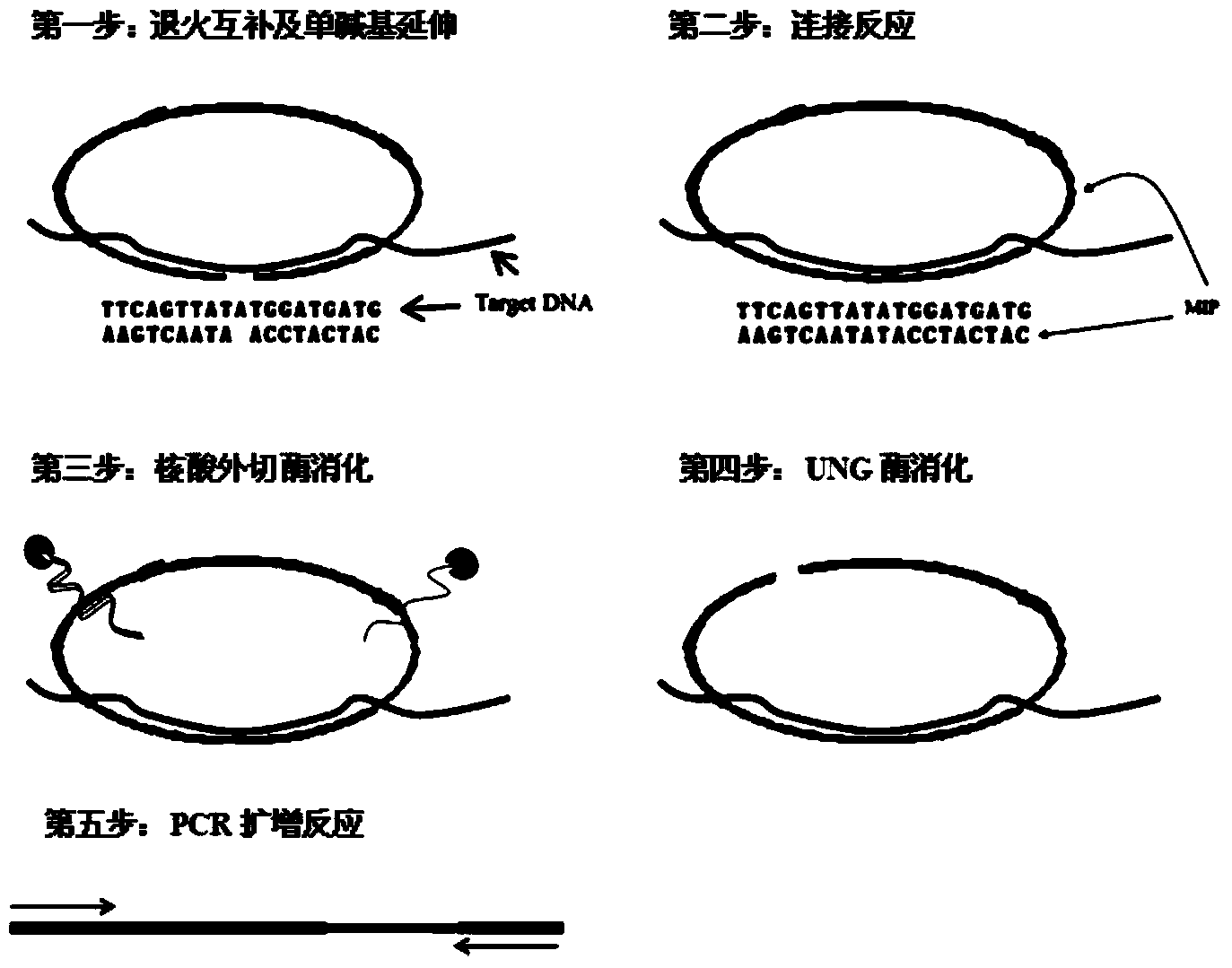
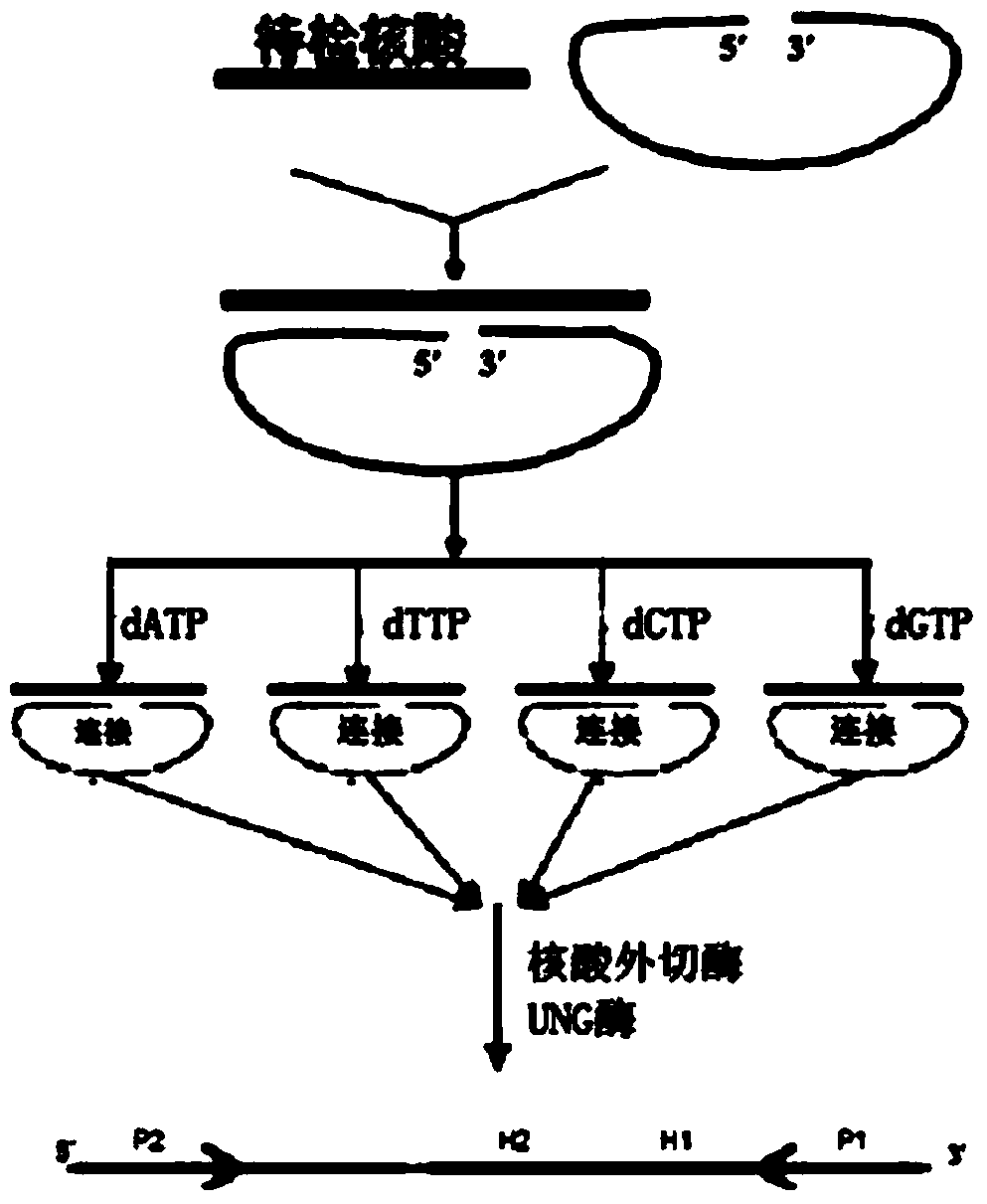
[0052] 1. Design of Molecular Inversion Probe:
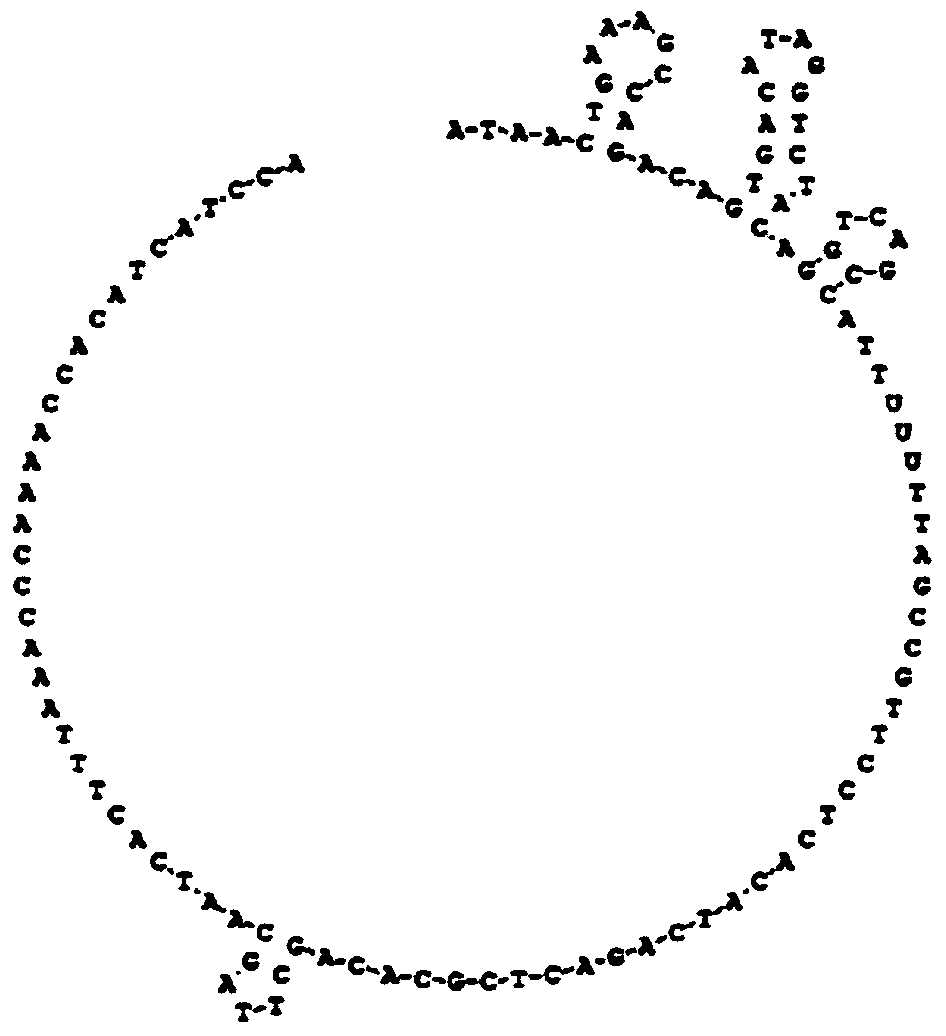
[0053] Aiming at the base sequences on both sides of the hepatitis B virus YVDD drug resistance site, a molecular inversion probe is designed, which consists of 6 parts: 2 complementary sequences of the target gene (gene complementary sequence A and gene complementary sequence B, respectively, as shown in SEQ ID NO:1, and SEQ ID NO:2), 1 UNG enzyme recognition site ("-TTUUUTT-"), 2 primer recognition sequences (respectively shown in SEQ ID NO:3 and SEQ ID NO: 4) and an unrelated sequence (shown in SEQ ID NO: 5) as the middle connecting sequence. There are 21 bases at the 5' end of the probe, and 19 bases at the 3' end to specifically recognize the base sequences on both sides of the YVDD drug resistance site. The secondary structure of the molecular inversion probe was analyzed using DNAMAN version6 software (Lynnon Biosoft, USA), and the irrelevant sequence in the middle was designed according to the principle of obtaining the...
Embodiment 2
[0080] Embodiment 2 Determination of Molecular Inversion Probe Detection Sensitivity
[0081] 1. The hepatitis B virus DNA was serially diluted to a concentration of 100nM, 10nM, 1nM, and 100pM.
[0082] 2. Apply the reaction system and optimized reaction conditions obtained in Example 1 to the determination of the detection sensitivity of the molecular inversion probe. That is, the molecular inversion probe was used to detect hepatitis B virus DNA at concentrations of 100 nM, 10 nM, 1 nM, and 100 pM, and the detection sensitivity was clarified.
[0083] The doubling dilution experiment of hepatitis B virus DNA shows that: the minimum template concentration that the molecular inversion probe can detect is 100pM (see Image 6 ).
Embodiment 3
[0084] Example 3 Detection of hepatitis B virus YVDD drug-resistant site clinical isolates
[0085] 1. Isolate hepatitis B virus DNA from the plasma of clinical hepatitis B patients, and use sequencing methods to analyze the YVDD drug resistance sites of clinical isolates of hepatitis B virus, and clarify the wild type of hepatitis B virus YVDD drug resistance sites. strains and mutants.
[0086] 2. Apply the reaction system and optimized reaction conditions obtained in Example 1 to the analysis of the molecular inversion probe to the YVDD drug resistance site of the clinical isolate of hepatitis B virus. By comparing the results of molecular inversion probes and sequencing methods on the analysis of YVDD drug resistance loci in clinical isolates of hepatitis B virus, the accuracy of molecular inversion probes in the detection of clinical isolates was clarified.
[0087] The experiment of molecular inversion probe used in the detection of clinical isolates shows that the mole...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com