Method, primers and probe for detecting relative expression quantity of 11q23/MLL fusion genes
A fusion gene and MLL-AF4 technology, applied in the field of life sciences and biology, can solve the problems that are not suitable for the detection of rare and unknown abnormalities, the inability to interpret small translocations, and the high requirements of the experimental environment, so as to improve the accuracy and detection efficiency , comprehensive diagnostic guidance, good pollution control effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0075] It is used to assist the clinical application of 11q23 / MLL series fusion genes MLL-AF4, MLL-AF6, MLL-AF9 and MLL in patients with ALL (acute lymphoblastic leukemia), AML (acute myeloid leukemia) and MDS (myelodysplastic syndrome) - A kit for detecting the relative expression level of ENL4mRNA, including:
[0076] Red blood cell lysate;
[0077] Trizol;
[0078] Chloroform;
[0080] Detection system PCR reaction solution: ReverTra Ace qPCR RT Kit (TOYOBO); THNDERBIRD Probe qPCR Mix (2×).
[0081] Primer and probe combinations are divided into MLL-AF4\MLL-AF6\MLL-AF9\MLL-ENL. in:
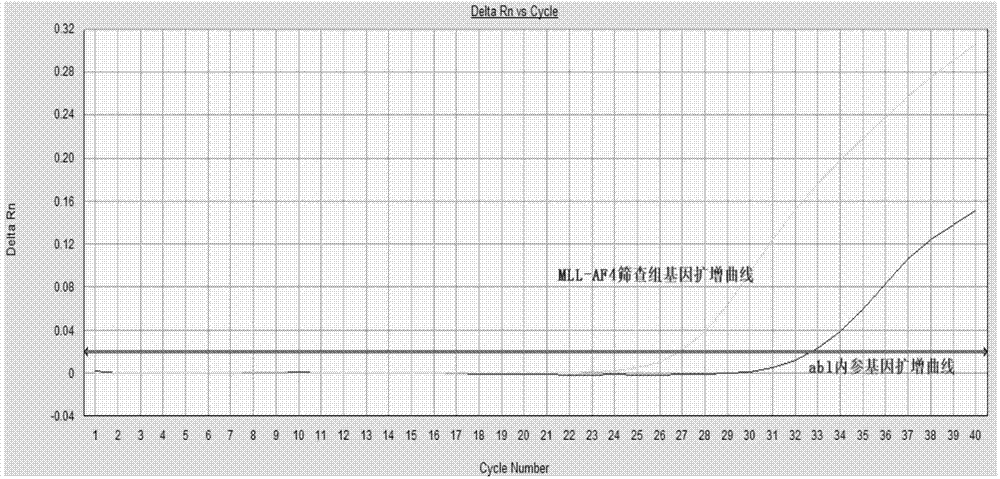
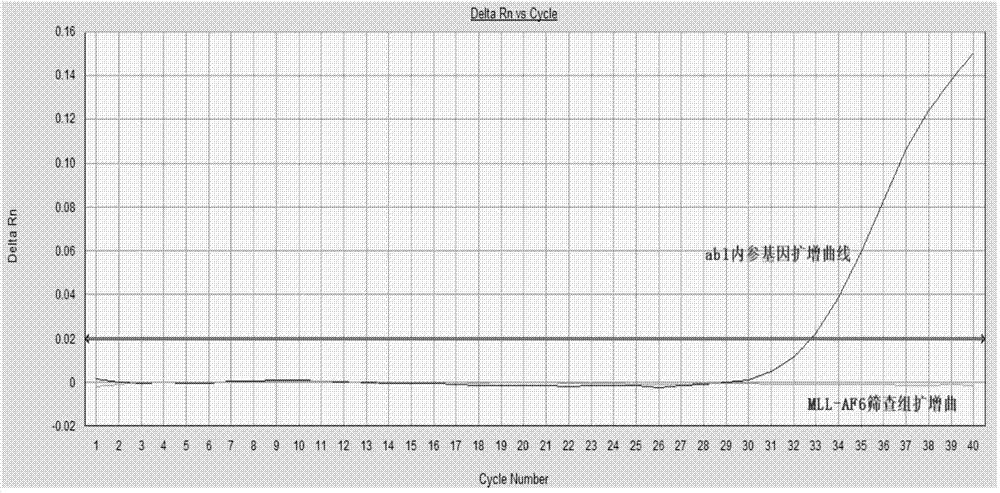
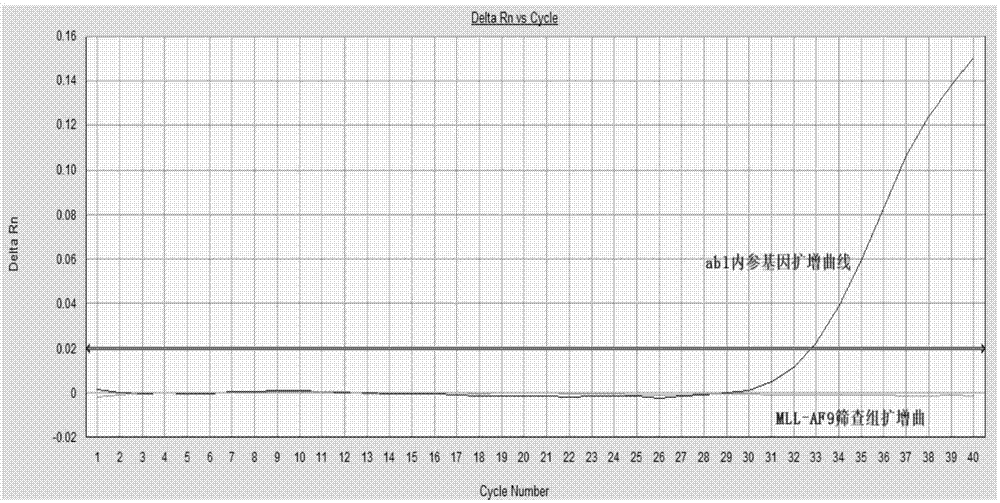
[0082]Screening group for detection of 4 fusion types of the target gene MLL-AF4: 0.8uM for the upstream and downstream primers, 0.4uM for the probe. The upstream primers are MLL-F1 and MLL-F2, the downstream primers are AF4-R1 and AF4-R2, and the probe follows the upstream primers, that is, the combination of P1 and MLL-F1, and the combination of P2 and MLL-F2...
Embodiment 2
[0118] Detection operation process:
[0119] (1) Extract tissue RNA from blood: Add 1ml of erythrocyte lysate to a clean 1.5ml centrifuge tube, take 0.5ml of anticoagulated blood and mix well. Let stand at room temperature for 10 minutes; centrifuge at 5000rpm for 5min, discard the supernatant, and collect the cells at the bottom; add 0.5ml red blood cell lysate again, centrifuge at 5000rpm for 5min, discard the supernatant, and collect the cells at the bottom; add 1ml Trizol to the cells, and pipette repeatedly until the precipitation is complete Dissolve, let stand at room temperature for 5 minutes; add 0.2ml chloroform, shake evenly; centrifuge at 14000rpm at 4°C for 10 minutes, absorb the supernatant layer and transfer to another new centrifuge tube; add an equal volume of isopropanol, mix well up and down, and let stand at room temperature for 10 minutes Centrifuge at 14000rpm at 4°C for 10min, discard the supernatant, add 1ml of 75% ethanol, wash the tube wall upside dow...
Embodiment 3
[0139] Take 20 samples each of ALL, AML, and MDS patients submitted for inspection, extract tissue RNA according to the method described in Example 2, and reverse-transcribe the extracted tissue RNA into cDNA, prepare and detect the PCR reaction solution of the detection system.
[0140] Each sample was reverse-transcribed into cDNA, and 2 μl of cDNA sample was added to the detection system PCR reaction solution. At the same time, make positive, negative, blank control, internal reference gene / target gene standard curve each. Each sample has 2 replicates, 1 positive control, 1 negative control and 1 blank control. The detection time is 100 minutes.
[0141] Experimental result of the present invention and △ △ CT The results of the method were compared to determine the accuracy of the sample detection. The results are shown in Table 1, Table 2 and Table 3:
[0142] Table 1: Test results of 20 ALL patients
[0143]
[0144] Table 2: Test results of 20 AML patients
[01...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com