Method for increasing yield of alpha-oxoglutarate produced through whole-cell transformation
A technology of whole cell conversion and ketoglutarate, applied in the direction of microorganism-based methods, biochemical equipment and methods, botany equipment and methods, etc., can solve the problems of pollution, low yield, high cost, etc., and achieve increased production Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
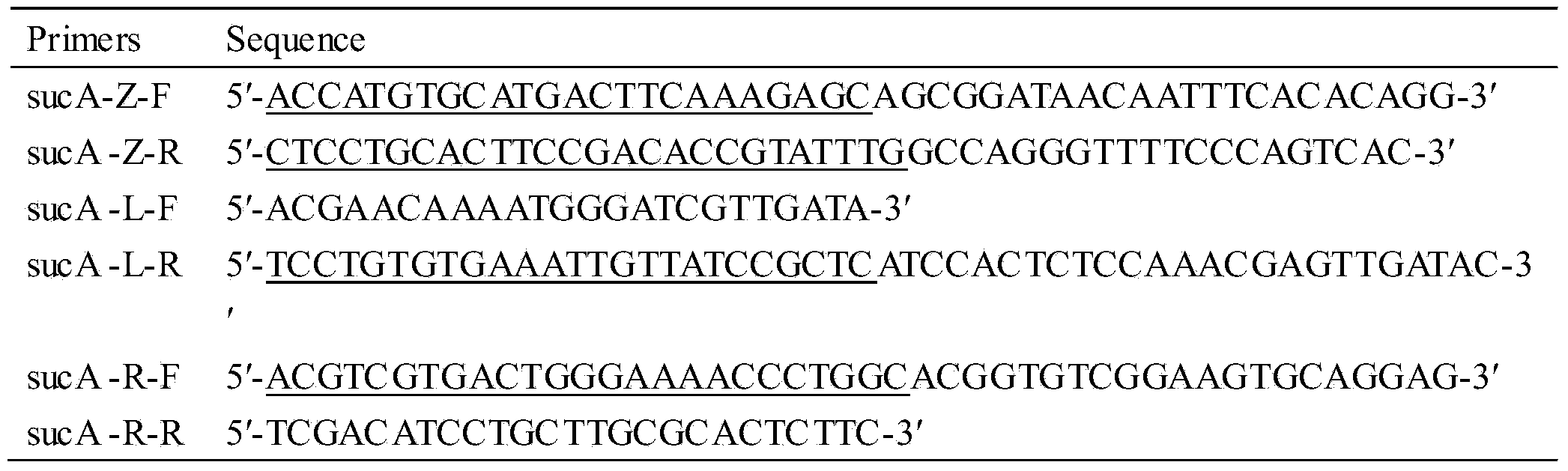
[0018] The knockout of embodiment 1 sucA gene
[0019] In a previous study, expression of an L-amino acid deaminase gene (SEQ ID NO.1) engineered by error-prone PCR or site-directed saturation mutagenesis in Bacillus subtilis improved whole-cell conversion of L-glutamate to α-ketopentamate Diacid conversion rate.
[0020]
[0021]
[0022] Using this recombinant Bacillus subtilis genomic DNA as a template, using sucA-L-F and sucA-L-R as primers, PCR amplifies about 1000 bp upstream of the sucA gene, and uses sucA-R-F and sucA-R-R as primers, PCR amplifies about 1000 bp downstream of the sucA gene . Using p7Z6 as a template and sucA-Z-F and sucA-Z-R as primers, the resistance marker fragment Lox71-zeo-lox66 was amplified about 1000bp. Fusion PCR was performed on these three fragments. Fusion PCR fragments were transformed into Bacillus subtilis, homologous recombination, and zeor transformants were screened. Transformation of zeo with thermosensitive plasmid pTSC r T...
Embodiment 2
[0026] Preparation of embodiment 2 whole cell catalyst and whole cell transformation process
[0027] The original Bacillus subtilis in Example 1 and the recombinant Bacillus subtilis after knocking out sucA were inoculated into the seed medium (chloramphenicol 10 mg / L), and cultivated overnight at 37°C and 200 rpm. Fermentation was carried out in a 3L NBS fermenter, 1% inoculum was added to 1.8L fermentation medium, the stirring speed, ventilation and temperature were 400rpm, 1.0vvm and 28°C, respectively, when OD 600 When 0.6 was reached, 0.4 mM IPTG was added to induce the expression of L-amino acid deaminase. After 5 hours of induction, centrifuge at 8,000 rpm for 10 minutes at low temperature, collect the bacterial cells, and wash the bacterial cells twice with 20 mM Tris-HCl (pH 8.0) buffer solution. The whole cell transformation system is: L-glutamic acid 15g / L, whole cell catalyst 20.0g / L, the reaction is carried out in 20mM Tris-HCl (pH8.0), 37°C, 200rpm transformati...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com