Method, kit and primers for detecting of species of Salmo
A technology for salmon and species, applied in the field of fish detection, can solve the problems of no damage to the fish body, complex operation, strong professionalism, etc., to ensure accuracy and uniqueness, strong repeatability, and high specificity of detection results Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
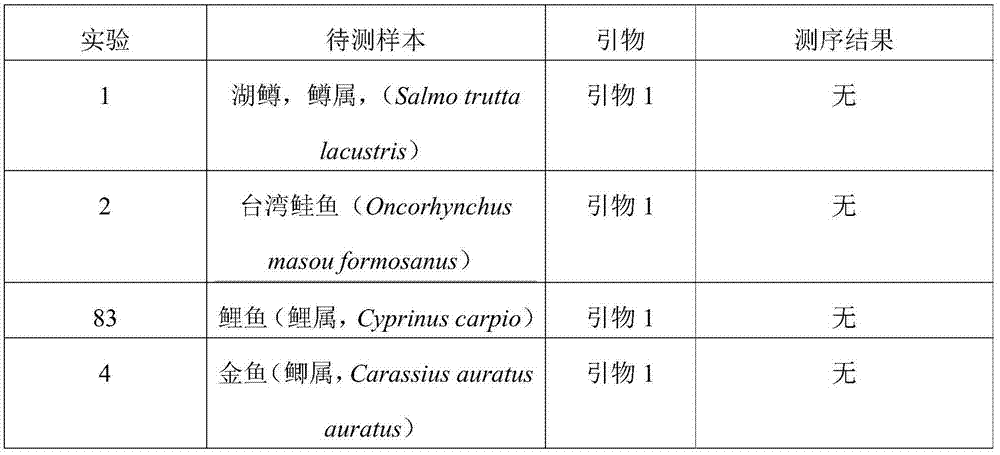
[0038] Embodiment 1 detects the design and synthesis of salmon primers
[0039] 1. According to the official website of NCBI (National Center for Biotechnology Information), search for a total of 445 COI genes of three species of salmon Salmosalar, Oncorhynchus mykiss and Oncorhynchus keta, listed as follows:
[0040] The cytochrome oxidase subunit I (COI) gene of salmon is as follows:
[0041] FJ999166 FJ998966 FJ998729
[0042] FJ999161 FJ998953 FJ998718
[0043] FJ999155 FJ998950 FJ998712
[0044] FJ999148 FJ998803 EU525057
[0045] FJ999131 FJ998799 HQ712702
[0046] FJ999122 FJ998788 HQ611128
[0047] FJ999118 FJ998777 GU440432
[0048] FJ999109 FJ998767 FJ918927
[0049] FJ998992 FJ998756 EF609449
[0050] FJ998986 FJ998745 FJ999539
[0051] FJ998973 FJ998739 FJ999511
[0052] FJ999508 JN007787 EU524353
[0053] FJ999503 JN007784 EU524349
[0054] FJ999485 JN007779 GU324184
[0055] FJ999472 HQ961026 HQ339988
[0056] FJ999469 HQ960962 EU752177
[0057] FJ...
Embodiment 2
[0077] Embodiment 2 detects salmon species
[0078] 1. Extract genomic DNA from the sample
[0079] Take 1g of commercially available salmon meat sample, add liquid nitrogen to grind for 10min, take 0.1g of the crushed sample and add it to a 2ml EP tube, add 1ml of cell lysate, 20μl of proteinase K, and mix well;
[0080] Bath in a constant temperature water bath at 62°C for 20 minutes, centrifuge at 12,000 rpm for 5 minutes in a desktop centrifuge, take the supernatant and carefully transfer it to another centrifuge tube, add 2 times the volume of isopropanol, invert and mix well and you can see filaments, use a 100μl tip pick out, dry
[0081] Add 200 μl TE to rewash and dissolve, add an equal amount of phenol: chloroform: isoamyl alcohol (25:24:1) to shake and mix, and centrifuge at 12,000 rpm for 5 min;
[0082] Take the upper layer solution to another tube, add an equal volume of chloroform:isoamyl alcohol (24:1), shake and mix, and centrifuge at 12000rpm for 5min;
[...
Embodiment 3
[0094] Embodiment 3 detects salmon species
[0095] 1. Extract genomic DNA from the sample
[0096] A salmon tissue sample was taken, and the DNA of the tissue sample was extracted according to the sample gene DNA group extraction method described in Example 2.
[0097] 2. PCR method to amplify the target gene
[0098] (1) PCR reaction
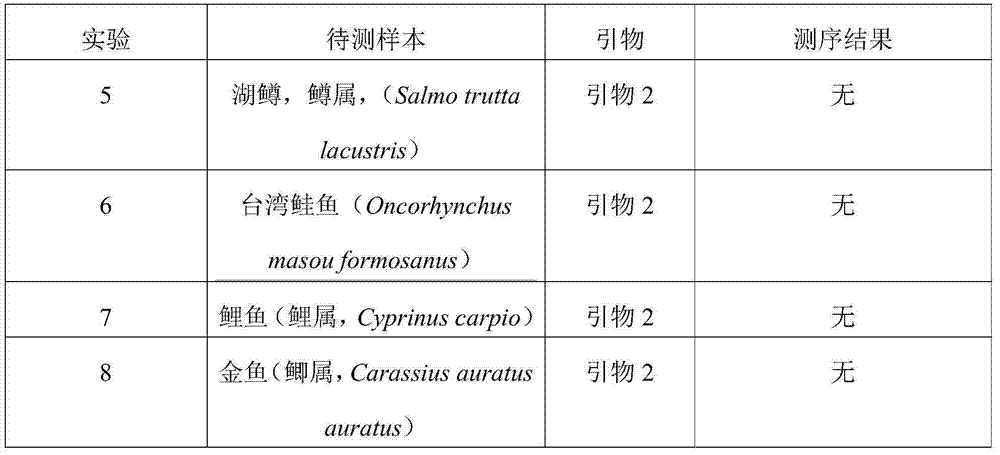
[0099] a. Primers: Take the primer 2 obtained in Example 1, and the forward and reverse primer sequences in the primer 2 are respectively shown in SEQ ID NO:3 and SEQ ID NO:4 in the sequence listing. According to the given concentration of the synthetic primers, a solution with a concentration of 10 μmol / L was prepared for use.
[0100] b. Mg in the PCR reaction system 2+ The concentration is 15mmol / L, the dNTP concentration is 0.3mmol / L, the amplification primer concentration is 0.3μM, the Taq enzyme concentration is 0.1U / μL, and the DNA template concentration is 100ng / μL.
[0101] c. The PCR amplification program is (1) pre-denaturation...
PUM
| Property | Measurement | Unit |
|---|---|---|
| melting point | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com