Efficient method for in-vitro evolution of serine hydroxymethyltransferase
A technology of serine hydroxymethyl and in vitro evolution, applied in the direction of transferase, introduction of foreign genetic material using vectors, recombinant DNA technology, etc., can solve the problems of only point mutation, sequence homology, low mutation rate, etc., and reach the site Multiple, simple operation, high frequency effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Start with Escherichia coli str.K-12substr.MG1655, and follow the steps below:
[0036] 1. Cultivate Escherichia coli in the following 50ml medium (g / L): peptone 10, yeast extract 5, sodium chloride 10, pH 7.2, and shake overnight at 37°C. The culture broth was collected, filtered with suction and dried with filter paper, and the genomic DNA was extracted with the kit and stored at -20°C. The serine hydroxymethyltransferase gene was used as the target gene, and the genomic DNA was used as the template. Taq DNA polymerase was used for PCR amplification and purification. The target fragment was 1254 bp in length. Design primers as follows:
[0037] Primer-F5': CGCGGATCCATGTTAAAGCGTG
[0038] Primer-R5': GGCAAGCTTTTATGCGTAAACC
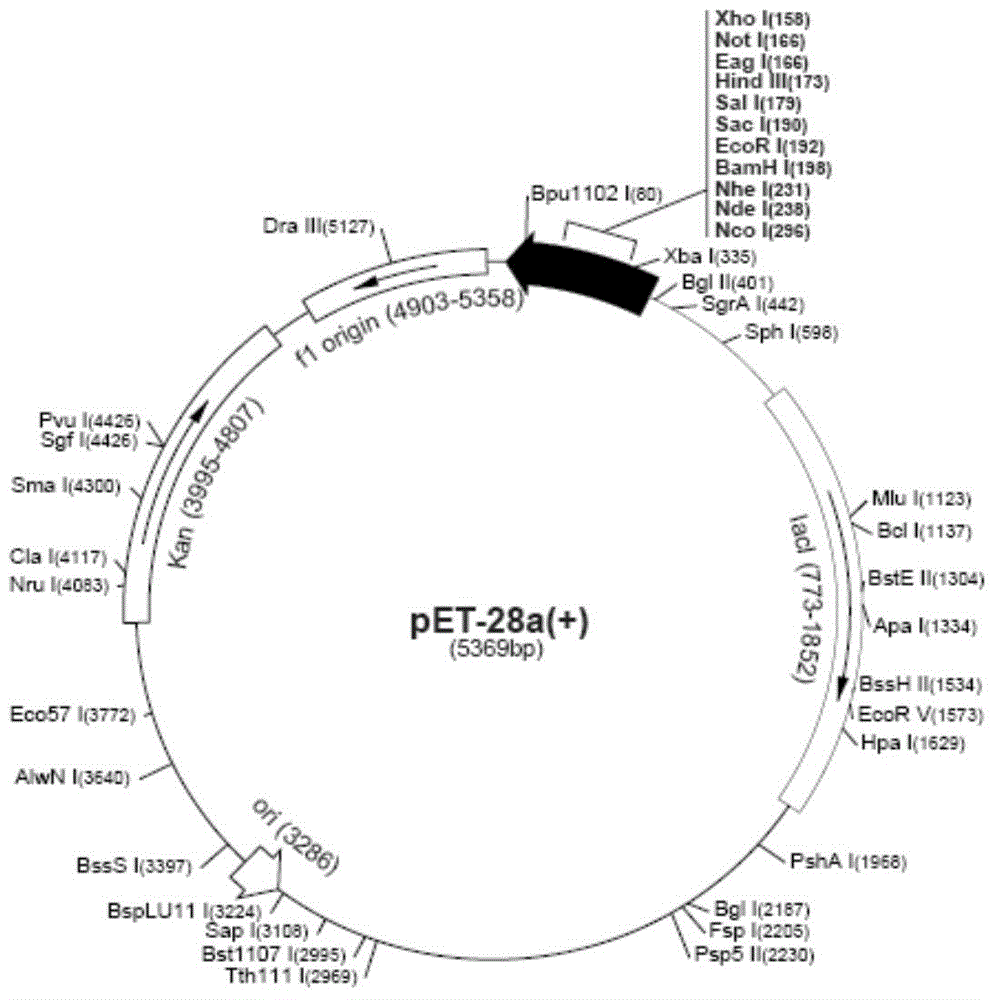
[0039] 2. After the purified PCR product is digested with BamHI / HindIII double enzymes, it is separated and purified by conventional agarose gel electrophoresis. The recovered product is placed in a sterile 96-well plate, 20μl per well, frozen at -20°C f...
Embodiment 2
[0044] Start with the Bacillus subtilis (ACCC10075) strain, and follow the steps below:
[0045] 1. Cultivate Bacillus subtilis in the following 50ml medium (g / L): peptone 10, beef extract 3, sodium chloride 5, pH 7.2, 37°C shaker overnight. The culture broth was collected, filtered with suction and dried with filter paper, and the genomic DNA was extracted with the kit and stored at -20°C. Taking serine hydroxymethyltransferase gene as the target gene, using genomic DNA as template, PCR amplification and purification with TaqDNA polymerase, the target fragment length is 1248bp. Design primers as follows:
[0046] Primer-F5': CGCGGATCCATGA AACATTTACC
[0047] Primer-R5': GGCCTCGAGTTAATAATCTAATTC
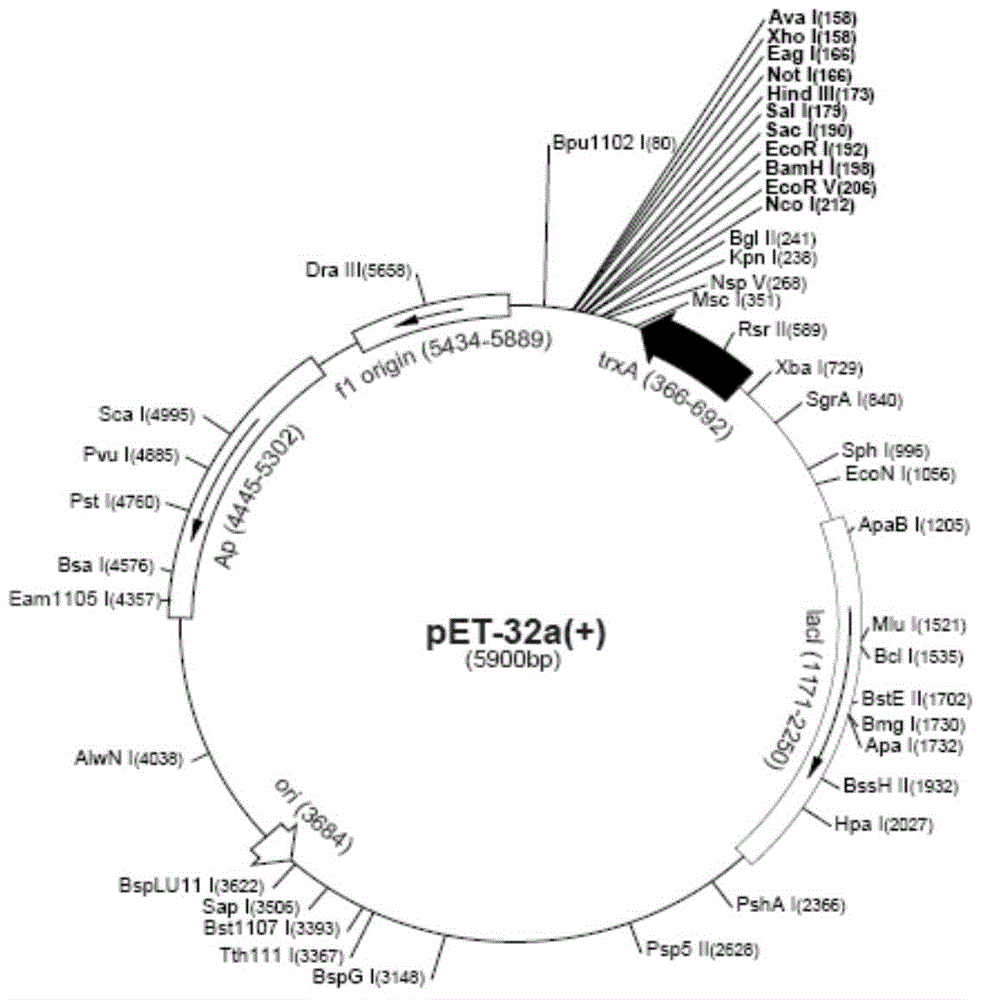
[0048] 2. After the purified PCR product is digested with BamHI / XhoI double enzyme, conventional agarose gel electrophoresis is separated, purified and recovered, and the recovered product is placed in a sterile 96-well plate with about 20μl per well. After freezing at -20°C for 30 minutes...
Embodiment 3
[0053] Start with Flavobacterium columnare ATCC49512 and follow the steps below:
[0054] 1. Cultivate Flavobacterium in the following 50mL medium (g / L): glycerol 10, peptone 10, yeast extract 1.5, sodium chloride 3, K 2 HPO 4 .12H 2 O4.5MgSO 4 .7H 2 O, natural pH, 37°C shaker overnight. The culture broth was collected, filtered with suction and dried with filter paper, and the genomic DNA was extracted with the kit and stored at -20°C. Taking serine hydroxymethyltransferase gene as the target gene, using genomic DNA as template, PCR amplification and purification with Taq DNA polymerase, the target fragment length is 1275bp. Design primers as follows:
[0055] Primer-F5': ATAGGATCCATGTTACG CGACG
[0056] Primer-R5': GGCCTCGAGTTAAAAAACGAAC
[0057] 2. After the purified PCR product is digested with BamHI / XhoI double enzymes, conventional agarose gel electrophoresis is used to separate, purify and recover. The recovered product is placed in a sterile 96-well plate, about 25μl per wel...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com