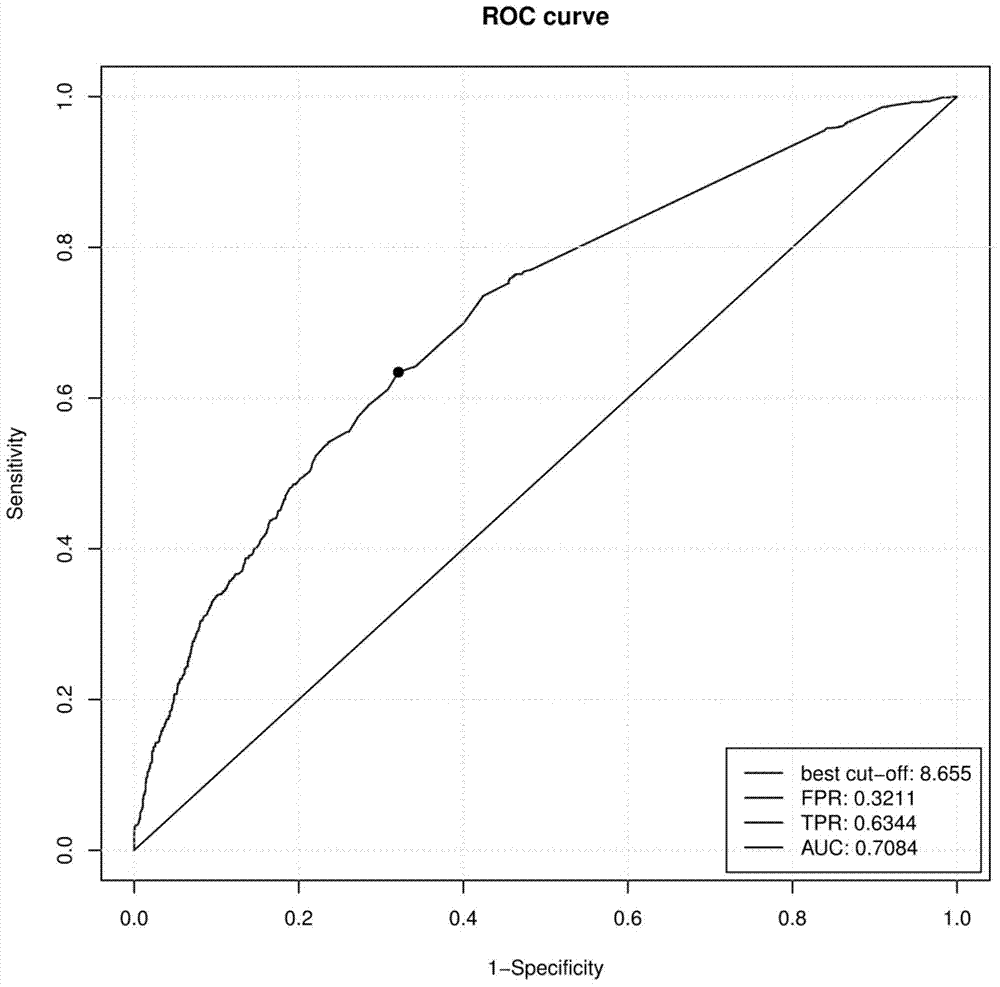
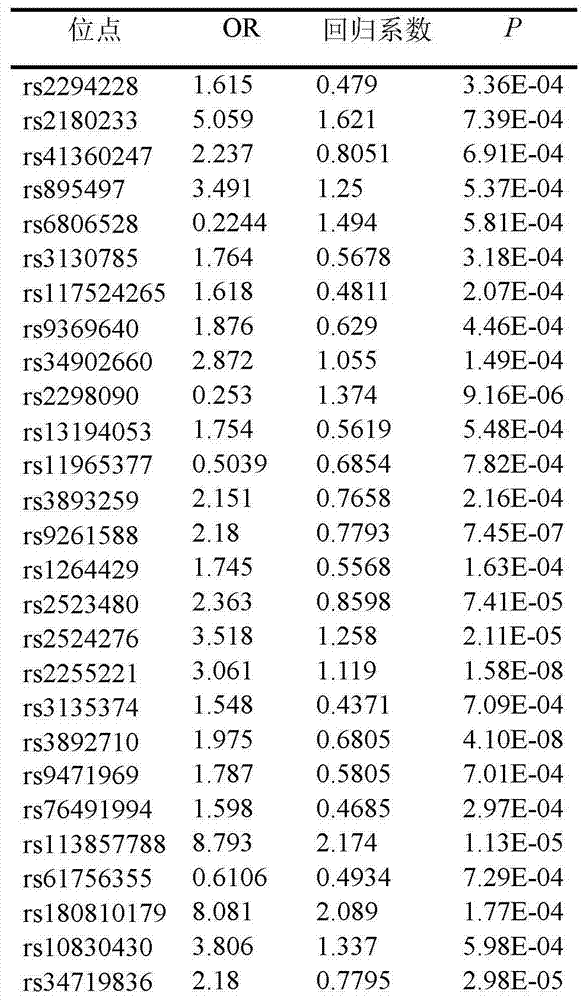
Low-frequency SNV (single nucleotide variant) marker related to assisted diagnosis of NOA (non-obstructive azoospermia) with unknown clinical reasons and application thereof
An auxiliary diagnosis and marker technology, applied in the fields of genetic engineering and reproductive medicine
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0069] The collection of embodiment 1 sample and the arrangement of sample data
[0070] The inventor collected a large number of blood samples of NOA patients from the Reproductive Medicine Center of Nanjing Medical University from April 2005 to January 2013. After sorting out the sample data, the inventor selected 5560 samples that met the following criteria. Experimental samples for microarray scanning and Sequenom MassARRAY genotyping of a single genetic marker:
[0071] 1. Repeated semen quality test / testicular biopsy, diagnosed as azoospermia;
[0072] 2. The semen of one ejaculation was centrifuged without sperm, and the cause of obstruction was ruled out;
[0073] 3. Normal sexual function; patients with known etiologies such as specific history of cryptorchidism, history of blood trauma, orchitis, vasectomy obstruction, vasectomy, polychromosomal abnormalities, and Y chromosome azoospermia factor microdeletion were excluded;
[0074] 4. Healthy male controls matched...
Embodiment 2
[0076] Example 2 Whole Exon Region Scanning of Low-Frequency SNVs in Peripheral Blood DNA
[0077] Among the above-mentioned eligible 960 patients with NOA of unknown clinical cause and 1800 healthy male controls, the two groups were age-matched. These two groups of people were passed through Illumina Human Exome BeadChip (Illumina Inc., San Diego, CA) chip detection obtained relevant results. The specific steps are:
[0078] 1. Add hemolysis reagent (i.e. lysate, 40 parts) to the peripheral blood stored in the 2ml cryopreservation tube. Dilute the TrisHcl solution to 2000ml, the same below), turn it upside down and mix it completely.
[0079] 2. Removal of red blood cells: Fill the 5ml centrifuge tube to 4ml with hemolysis reagent, mix by inverting, centrifuge at 4000rpm for 10 minutes, and discard the supernatant. Add 4ml of hemolysis reagent to the precipitate, invert and wash again, centrifuge at 4000rpm for 10 minutes, and discard the supernatant.
[0080] 3. Extrac...
Embodiment 3
[0090] Example 3 Sequenom MassARRAY Genotyping
[0091] The low-frequency and high-penetrance genetic markers found to be associated with the onset of NOA with clinically unknown causes were detected in another 1400 cases of NOA with clinically unknown causes and 1400 healthy male controls. The specific steps are as follows:
[0092] 1. Add the hemolysis reagent to the peripheral blood stored in the 2ml cryopreservation tube, mix it upside down and transfer it completely.
[0093] 2. Removal of red blood cells: Fill the 5ml centrifuge tube to 4ml with hemolysis reagent, mix by inverting, centrifuge at 4000rpm for 10 minutes, and discard the supernatant. Add 4ml of hemolysis reagent to the precipitate, invert and wash again, centrifuge at 4000rpm for 10 minutes, and discard the supernatant.
[0094] 3. Extract DNA: Add 1ml extract solution to the precipitate (each 300ml contains 122.5ml 0.2M sodium chloride, 14.4ml 0.5M ethylenediaminetetraacetic acid, 15ml 10% sodium dodecyl ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com