Phospholipase C
A technology of phospholipase and enzyme solution, applied in the field of enzyme degumming, can solve the problems of high enzyme activity, inability to completely remove phospholipids, and inability to find pH range, etc., and achieve the effect of cost saving and wide hydrolysis substrate characteristics.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0151] Embodiment 1, PFSMC expression and purification
[0152] The sequence shown in SEQ ID NO: 2 (its coding sequence is shown in SEQ ID NO: 1) was synthesized by Sangon Bioengineering (Shanghai) Co., Ltd., and cloned in the expression vector pET24a(+).
[0153] The pET24a (+) with this nucleotide sequence is transformed into Escherichia coli BL21 (DE3) to induce expression (refer to "Molecular Cloning Experiment Guide", the third edition, Science Press 2002, [US] J. Sambrook Written by D.W Russell, translated by Huang Peitang, etc.). The specific process is as follows:
[0154] Pick positive transformants and culture overnight at 37°C in LB medium, inoculate the seed culture solution into the expression medium at 1% inoculum size, and culture at 37°C, 220 rpm until OD600=0.6-0.8.
[0155] After cooling to 20°C, add IPTG to 0.1mM for induction, and express overnight at 20°C and 190rpm. After induction of expression, the cells were collected by centrifugation. Resuspend t...
Embodiment 2
[0158] Embodiment 2, PFSMC substrate preference
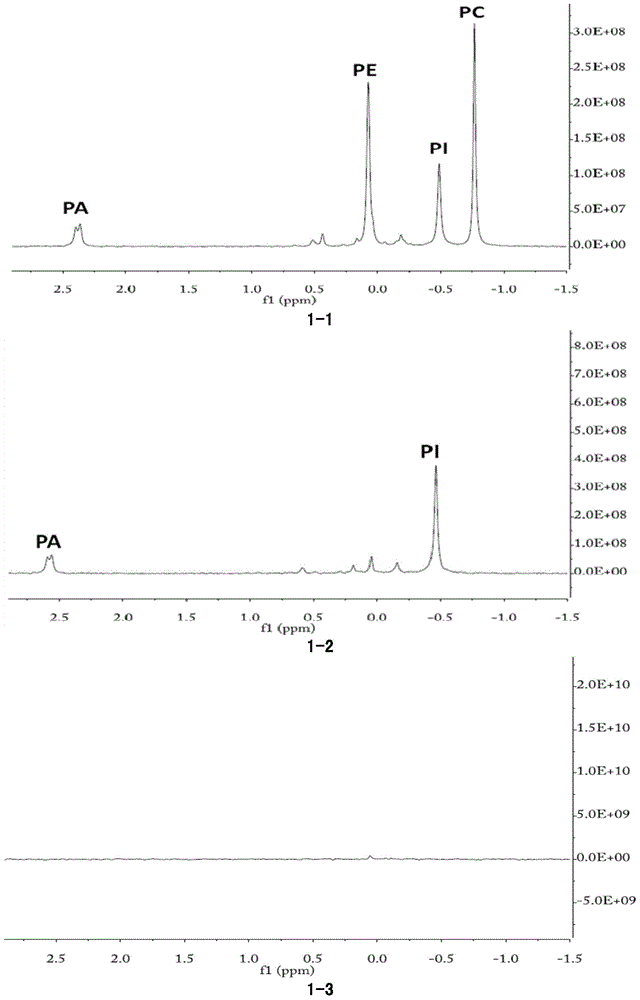
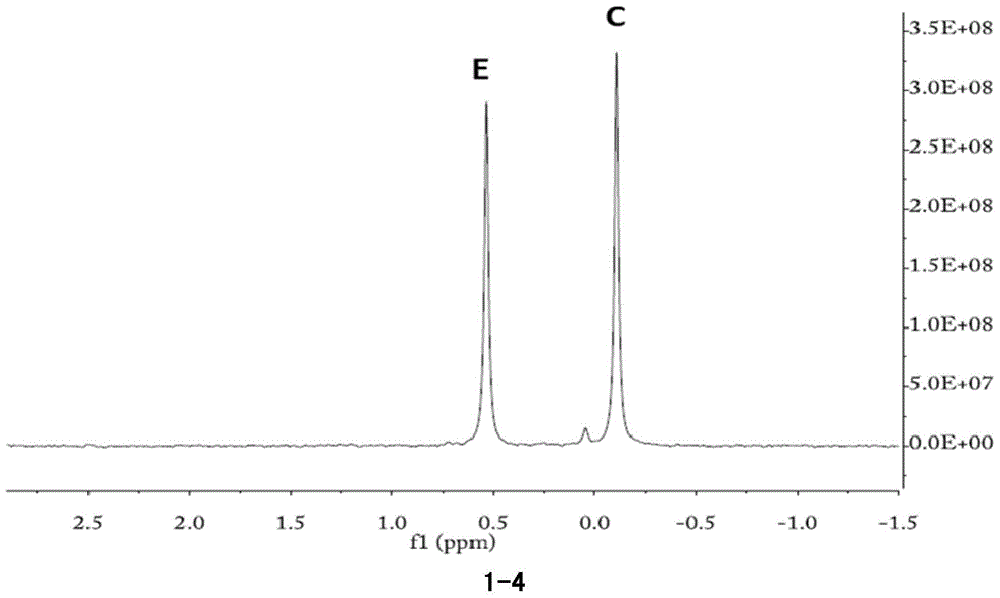
[0159] Mix phospholipid substrate (15.75g / L soybean powder phospholipid (a mixture of phosphatidylcholine, phosphatidylinositol, phosphatidylserine, phosphatidylethanolamine phosphatidic acid, etc.), 5mM CaCl into 10ml 2 ), add 0.5ml PFSMC (2.9mg / ml) to another tube of 10ml phospholipid mixed substrate, add 0.5ml water as a control, react at 35℃, 200rpm for 18 hours, add 10ml chloroform, shake and mix, and centrifuge at 10,000g for 10 minutes , the solution is divided into upper and lower layers.
[0160] Take 1ml of the upper aqueous phase, lyophilize and resuspend in 1ml of heavy water. Take 1ml of the lower chloroform phase, dry it in an ultra-clean bench, resuspend in 3ml of NMR analysis buffer, and conduct 31P-NMR analysis on the two phases respectively.
[0161] The NMR results of the phospholipid substrate chloroform phase added with water are shown in Picture 1-1 , Visible, phospholipid substrate mainly contains thr...
Embodiment 3
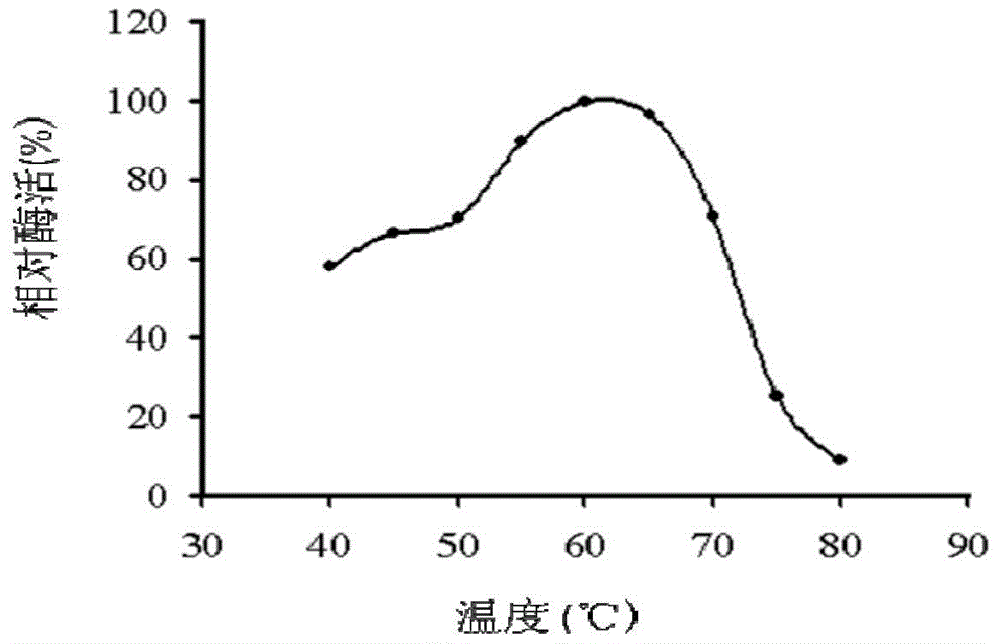
[0163] Part enzymatic properties of embodiment 3, PFSMC
[0164] In a 200 μL reaction system, containing soybean powder phospholipids 0.5% (w / v), buffer system (25mM Tris-Cl, pH7.5), 5mM CaCl 2 , add phospholipase PFSMC 20μl (0.097mg / ml), react for 30min, add 200μL of chloroform, shake and mix for 30s, carry out centrifugation at 12,000rpm for 1min, take 80μL supernatant and add it to the phospholipase reaction system, the final volume is 200μL, the system Also contains 50mM Tris-Cl (pH9.0), 10mM MgCl 2 , CIAP 10 U / μL. The reaction was carried out in a 37 °C water bath for 30 min. Add 740 μL of deionized water, 20 μL of 10% (w / v) ascorbic acid, and 40 μL of 2.5% (w / v) ammonium molybdate solution. Develop color at 37°C for 10 minutes. The solution after color development was taken for absorbance detection at 700nm. Combined with the standard curve detected by phosphomolybdenum blue, the data is analyzed and regressed, and after multiplying by the dilution factor, the activ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com